2ZVI
 
 | | Crystal structure of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | | Authors: | Tamura, H, Yadani, T, Kai, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2YRF
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis complexed with sulfate ion | | Descriptor: | Methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
1K1Y
 
 | | Crystal structure of thermococcus litoralis 4-alpha-glucanotransferase complexed with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-ALPHA-GLUCANOTRANSFERASE, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1X
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
2CW8
 
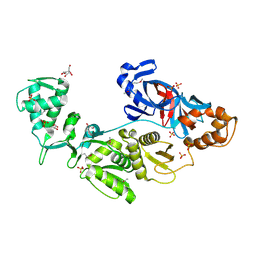 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2CW7
 
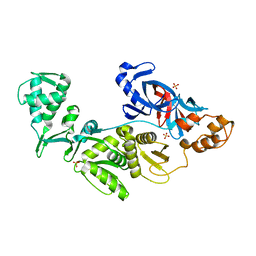 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
1K1W
 
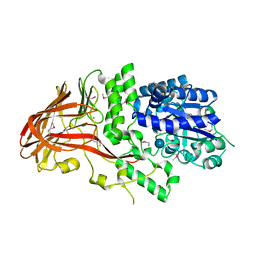 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
3B1J
 
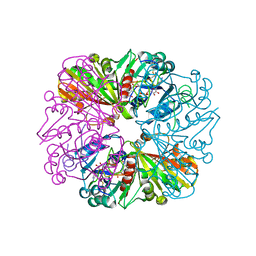 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with CP12 in the presence of copper from Synechococcus elongatus | | Descriptor: | COPPER (II) ION, CP12, Glyceraldehyde 3-phosphate dehydrogenase (NADP+), ... | | Authors: | Matsumura, H, Kai, A, Inoue, T. | | Deposit date: | 2011-07-04 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
3B20
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with NADfrom Synechococcus elongatus" | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Matsumura, H, Kai, A, Maeda, T, Inoue, T. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
3AXM
 
 | | Structure of rice Rubisco in complex with 6PG | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3AXK
 
 | | Structure of rice Rubisco in complex with NADP(H) | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Matsumura, H, Mizohata, E, Ishida, H, Kogami, A, Ueno, T, Makino, A, Inoue, T, Yokota, A, Mae, T, Kai, Y. | | Deposit date: | 2011-04-11 | | Release date: | 2012-04-11 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
3B1K
 
 | |
2ZA0
 
 | | Crystal structure of mouse glyoxalase I complexed with methyl-gerfelin | | Descriptor: | Glyoxalase I, ZINC ION, methyl 4-(2,3-dihydroxy-5-methylphenoxy)-2-hydroxy-6-methylbenzoate | | Authors: | Okumura, H, Kawatani, M, Osada, H. | | Deposit date: | 2007-09-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3WB1
 
 | | HcgB from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0488 | | Authors: | Tamura, H, Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2013-05-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the HcgB enzyme in [Fe]-hydrogenase-cofactor biosynthesis.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2DSN
 
 | | Crystal structure of T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Matsumura, H, Kai, Y. | | Deposit date: | 2006-07-03 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
3WRY
 
 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRX
 
 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZHP
 
 | | Crystal structure of bleomycin-binding protein from Streptoalloteichus hindustanus complexed with bleomycin derivative | | Descriptor: | Bleomycin resistance protein, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2008-02-06 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Affinity Relationship Study of Bleomycins and Shble protein Using a Chemical Array
To be Published
|
|
3ACL
 
 | | Crystal Structure of Human Pirin in complex with Triphenyl Compound | | Descriptor: | FE (II) ION, N-{[4-(benzyloxy)phenyl](methyl)-lambda~4~-sulfanylidene}-4-methylbenzenesulfonamide, Pirin | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2010-01-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule inhibitor shows that pirin regulates migration of melanoma cells
Nat.Chem.Biol., 6, 2010
|
|
3WRV
 
 | |
2Z5G
 
 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
5YAE
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 2.4 A resolution | | Descriptor: | ACETATE ION, Esterase, SULFATE ION | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-08-31 | | Release date: | 2017-12-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | Descriptor: | ACETATE ION, Esterase, GLYCEROL, ... | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-09-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
3WZJ
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-(6-(cyclohexylamino)-8-(((tetrahydro-2H-pyran-4-yl)methyl)amino)imidazo[1,2-b]pyridazin-3-yl)-N-cyclopropylbenzamide | | Descriptor: | 4-{6-(cyclohexylamino)-8-[(tetrahydro-2H-pyran-4-ylmethyl)amino]imidazo[1,2-b]pyridazin-3-yl}-N-cyclopropylbenzamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
3WYX
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
