1TFR
 
 | | RNASE H FROM BACTERIOPHAGE T4 | | Descriptor: | MAGNESIUM ION, T4 RNASE H | | Authors: | Mueser, T.C, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1996-04-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of bacteriophage T4 RNase H, a 5' to 3' RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins.
Cell(Cambridge,Mass.), 85, 1996
|
|
1C1K
 
 | | BACTERIOPHAGE T4 GENE 59 HELICASE ASSEMBLY PROTEIN | | Descriptor: | BPT4 GENE 59 HELICASE ASSEMBLY PROTEIN, CHLORIDE ION, IRIDIUM ION | | Authors: | Mueser, T.C, Jones, C.E, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1999-07-22 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 A resolution crystal structure reveals a novel alpha-helical two-domain fold.
J.Mol.Biol., 296, 2000
|
|
1G09
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 7.2 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
1G0B
 
 | | CARBONMONOXY LIGANDED EQUINE HEMOGLOBIN PH 8.5 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
1G08
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 5.0 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
1G0A
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 8.5 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
5TOQ
 
 | | High resolution crystal structure of AAT | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5TON
 
 | | Crystal structure of AAT H143L mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5TOR
 
 | | Crystal structure of AAT D222T mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5TOT
 
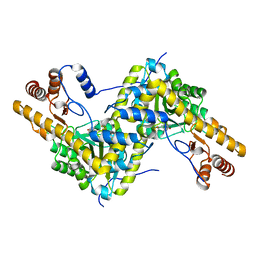 | |
5C6E
 
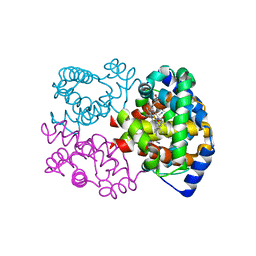 | | Joint X-ray/neutron structure of equine cyanomet hemoglobin in R state | | Descriptor: | CYANIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dajnowicz, S, Sean, S, Hanson, B.L, Fisher, S.Z, Langan, P, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2015-06-22 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the Bohr effect in hemoglobin: neutron structure of equine cyanomethemoglobin in the R state and comparison with human deoxyhemoglobin in the T state.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1QBZ
 
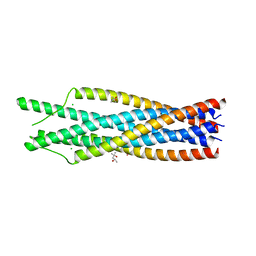 | | THE CRYSTAL STRUCTURE OF THE SIV GP41 ECTODOMAIN AT 1.47 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, MERCURY (II) ION, ... | | Authors: | Yang, Z.-N, Mueser, T.C, Kaufman, J, Stahl, S.J, Wingfield, P.T, Hyde, C.C. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution.
J.Struct.Biol., 126, 1999
|
|
2IHN
 
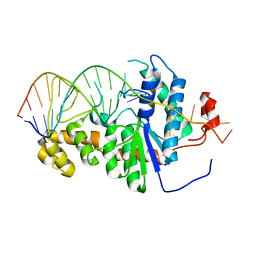 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
7TUR
 
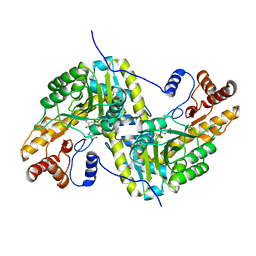 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
8EYP
 
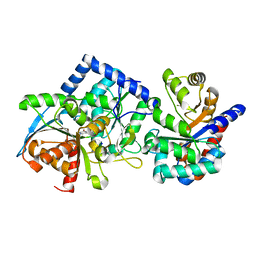 | | Joint X-ray/neutron structure of Salmonella typhimurium tryptophan synthase internal aldimine from microgravity-grown crystal | | Descriptor: | SODIUM ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Drago, V.N, Kovalevsky, A, Blakeley, M.P, Forsyth, V.T, Mueser, T.C. | | Deposit date: | 2022-10-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Neutron diffraction from a microgravity-grown crystal reveals the active site hydrogens of the internal aldimine form of tryptophan synthase.
Cell Rep Phys Sci, 5, 2024
|
|
8EZC
 
 | |
8EYS
 
 | |
7MT4
 
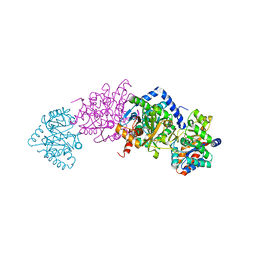 | | Crystal structure of tryptophan Synthase in complex with F9, NH4+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMMONIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT5
 
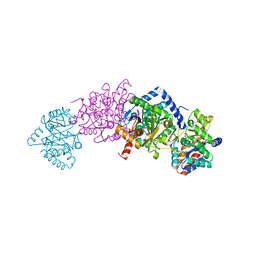 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, CESIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT6
 
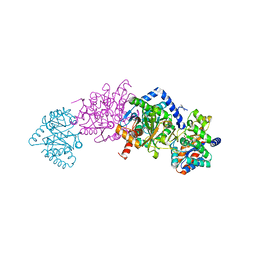 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, benzimidazole, pH7.8 - alpha aminoacrylate form - E(A-A)(BZI) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3RDE
 
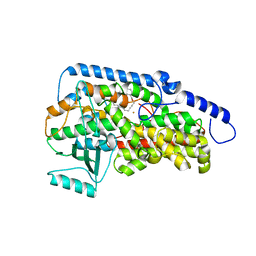 | | Crystal structure of the catalytic domain of porcine leukocyte 12-lipoxygenase | | Descriptor: | 3-{4-[(tridec-2-yn-1-yloxy)methyl]phenyl}propanoic acid, Arachidonate 12-lipoxygenase, 12S-type, ... | | Authors: | Funk, M.O, Xu, S, Marnett, L.J, Mueser, T.C. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis.
Structure, 20, 2012
|
|
3H8J
 
 | |
3H8S
 
 | |
3H8W
 
 | |
3H7I
 
 | | Structure of the metal-free D132N T4 RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Tomanicek, S.J, Mueser, T.C. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Additional Order Appears in the Absence of Metals in a FEN-1 protein: Structural Analysis of Magnesium Binding to Bacteriophage T4 RNaseH
To be Published
|
|
