8QJZ
 
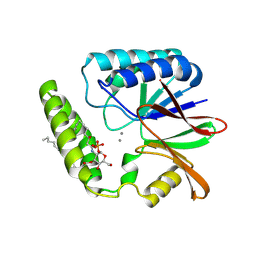 | | Crystal structure of E. coli LpxH in complex with lipid X | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QKA
 
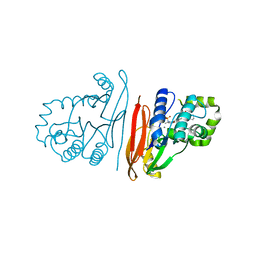 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1Z3W
 
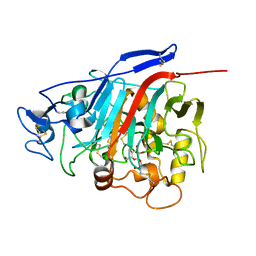 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Vasella, A, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3T
 
 | |
1Z3V
 
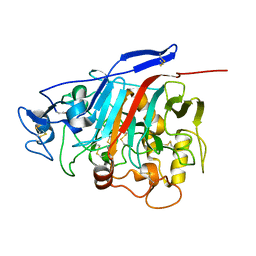 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Munoz, I.G, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
3CXU
 
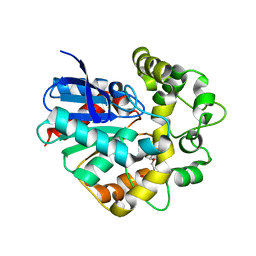 | | Structure of a Y149F mutant of epoxide hydrolase from Solanum tuberosum | | Descriptor: | Epoxide hydrolase, TETRAETHYLENE GLYCOL | | Authors: | Naworyta, A, Mowbray, S.L, Widersten, M, Thomaeus, A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability
Protein Sci., 17, 2008
|
|
1GQT
 
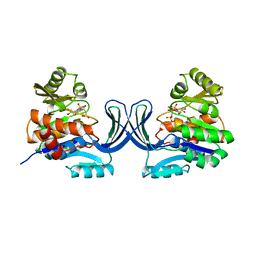 | |
4ACF
 
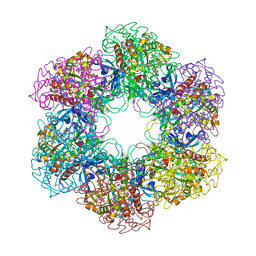 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE IN COMPLEX WITH IMIDAZOPYRIDINE INHIBITOR ((4-(6-BROMO-3-(BUTYLAMINO)IMIDAZO(1,2-A)PYRIDIN-2-YL)PHENOXY) ACETIC ACID) AND L-METHIONINE-S-SULFOXIMINE PHOSPHATE. | | Descriptor: | CHLORIDE ION, GLUTAMINE SYNTHETASE 1, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Nilsson, M.T, Mowbray, S.L. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, Biological Evaluation and X-Ray Crystallographic Studies of Imidazo(1,2-A)Pyridine-Based Mycobacterium Tuberculosis Glutamine Synthetase Inhibitors
Medchemcomm, 3, 2012
|
|
3ZXV
 
 | |
3ZXR
 
 | |
4Y6R
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC137, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CHLORIDE ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
4Y6S
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC134, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MANGANESE (II) ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
4Y67
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC176, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MANGANESE (II) ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
4Y6P
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC177, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CALCIUM ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
3CAI
 
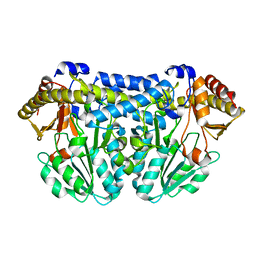 | | Crystal structure of Mycobacterium tuberculosis Rv3778c protein | | Descriptor: | POSSIBLE AMINOTRANSFERASE | | Authors: | Covarrubias, A.S, Larsson, A.M, Jones, T.A, Bergfors, T, Mowbray, S.L, Unge, T. | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical function studies of Mycobacterium tuberculosis essential protein Rv3778c
To be published
|
|
4Y9S
 
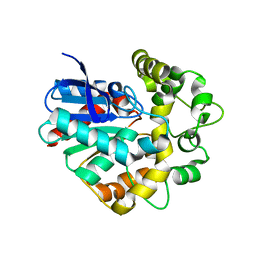 | |
2Z39
 
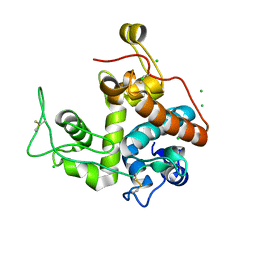 | |
2Z37
 
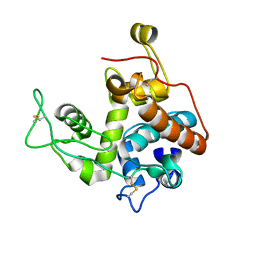 | |
2Z38
 
 | |
3G02
 
 | |
3G0I
 
 | |
2BES
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronohydroxamic acid. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism.
J.Biol.Chem., 280, 2005
|
|
3K7P
 
 | |
2BET
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronate. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONATE, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism
J.Biol.Chem., 280, 2005
|
|
3K7O
 
 | |
