6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
6YPI
 
 | | Structure of the engineered metallo-Diels-Alderase DA7 W16G,K58Q,L77R,T78R | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BENZOIC ACID, DA7 W16G,K58Q,L77R,T78R, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
5OJA
 
 | | Structure of MbQ | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJB
 
 | | Structure of MbQ NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJC
 
 | | Structure of MbQ2.1 NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
7BVT
 
 | | Crystal structure of cyclic alpha-maltosyl-1,6-maltose binding protein from Arthrobacter globiformis | | Descriptor: | Hypothetical sugar ABC-transporter sugar binding protein, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular analysis of cyclic alpha-maltosyl-(1→6)-maltose binding protein in the bacterial metabolic pathway.
Plos One, 15, 2020
|
|
5OJ9
 
 | | Structure of Mb NMH | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
6LII
 
 | | A quinone oxidoreductase | | Descriptor: | Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Hakoshima, T, Kim, S.-Y, Mori, T. | | Deposit date: | 2019-12-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into vesicle amine transport-1 (VAT-1) as a member of the NADPH-dependent quinone oxidoreductase family.
Sci Rep, 11, 2021
|
|
6KSV
 
 | | Crystal structure of SurE with D-Leu | | Descriptor: | Alpha/beta hydrolase, D-LEUCINE, S-(2-acetamidoethyl) (2R)-2-azanyl-4-methyl-pentanethioate, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2019-08-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Heterochiral coupling in non-ribosomal peptide macrolactamization
Nat Catal, 2020
|
|
6KSU
 
 | | Crystal structure of SurE | | Descriptor: | Alpha/beta hydrolase, MALONATE ION, SULFATE ION, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2019-08-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterochiral coupling in non-ribosomal peptide macrolactamization
Nat Catal, 2020
|
|
5B6K
 
 | |
5N81
 
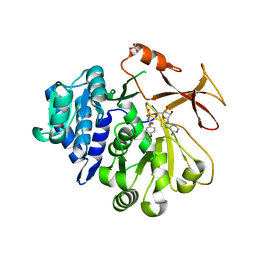 | | Crystal structure of an engineered TycA variant in complex with an O-propargyl-beta-Tyr-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.A, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
5N82
 
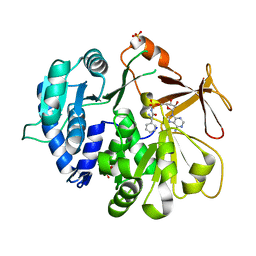 | | Crystal structure of an engineered TycA variant in complex with an beta-Phe-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.L, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
6LCD
 
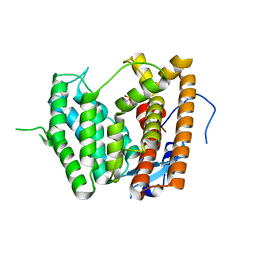 | |
6LCC
 
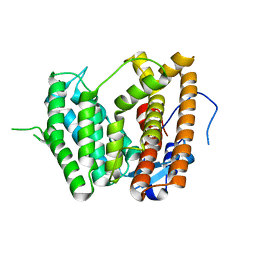 | |
1TF7
 
 | | Crystal Structure of Circadian Clock Protein KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC | | Authors: | Pattanayek, R, Wang, J, Mori, T, Xu, Y, Johnson, C.H, Egli, M. | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing a Circadian Clock Protein; Crystal Structure of KaiC and Functional Insights
Mol.Cell, 15, 2004
|
|
6A0L
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with maltose | | Descriptor: | Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
5ZXG
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, ligand-free form | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-05-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
1L5E
 
 | | The domain-swapped dimer of CV-N in solution | | Descriptor: | Cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1L5B
 
 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
6A0J
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with Cyclic alpha-maltosyl-(1-->6)-maltose | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
6A0K
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with panose | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
1LOM
 
 | | CYANOVIRIN-N DOUBLE MUTANT P51S S52P | | Descriptor: | CALCIUM ION, Cyanovirin-N, SULFATE ION | | Authors: | Botos, I, Mori, T, Cartner, L.K, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Domain-swapped structure of a mutant of cyanovirin-N.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
