5YRA
 
 | | Crystal Structure of Cypovirus Polyhedra R13A/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YRC
 
 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/E73C/Y83C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YR9
 
 | | Crystal Structure of Cypovirus Polyhedra R13A/E73C/Y83C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YR1
 
 | | Crystal Structure of Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
5YRD
 
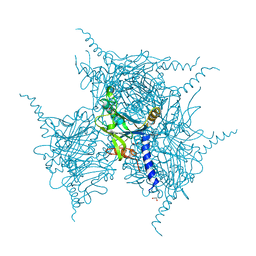 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
2ZID
 
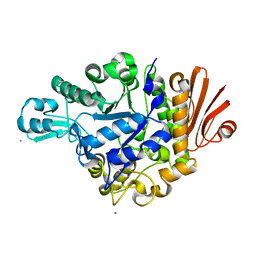 | | Crystal structure of dextran glucosidase E236Q complex with isomaltotriose | | Descriptor: | CALCIUM ION, Dextran glucosidase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
2ZIC
 
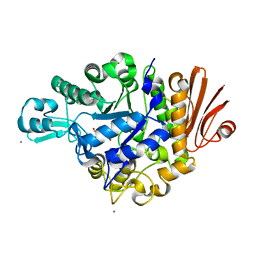 | | Crystal structure of Streptococcus mutans dextran glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Dextran glucosidase, ... | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
5YHA
 
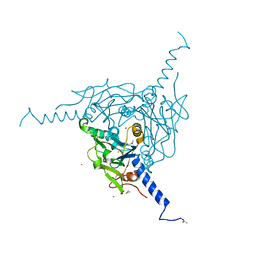 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
5YHB
 
 | | Crystal structure of Pd(allyl)/polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | PALLADIUM ION, Palladium(II) allyl complex, Polyhedrin | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
7E9X
 
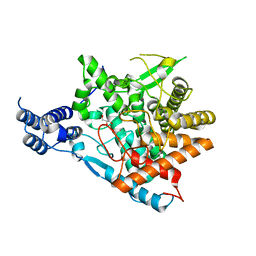 | | Trehalase of Arabidopsis thaliana acid mutant -D380A | | Descriptor: | GLYCEROL, Trehalase | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7E9U
 
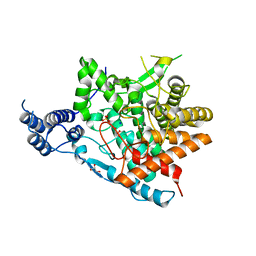 | | Trehalase of Arabidopsis thaliana | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7EAW
 
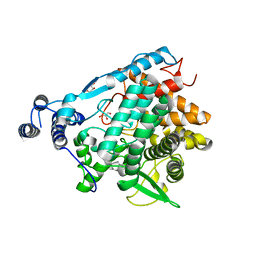 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
3VMP
 
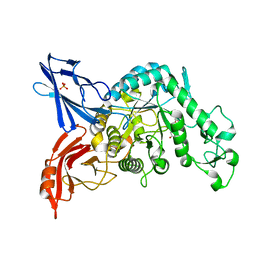 | | Crystal structure of dextranase from Streptococcus mutans in complex with 4,5-epoxypentyl alpha-D-glucopyranoside | | Descriptor: | 5-hydroxypentyl alpha-D-glucopyranoside, Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
3VMN
 
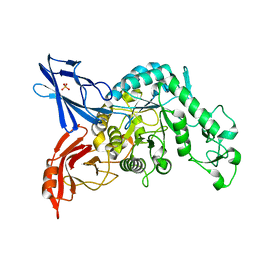 | | Crystal structure of dextranase from Streptococcus mutans | | Descriptor: | Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
3VMO
 
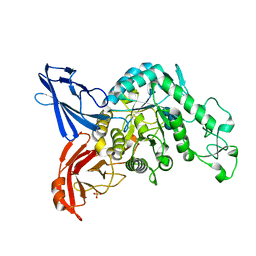 | | Crystal structure of dextranase from Streptococcus mutans in complex with isomaltotriose | | Descriptor: | Dextranase, PHOSPHATE ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
3WFA
 
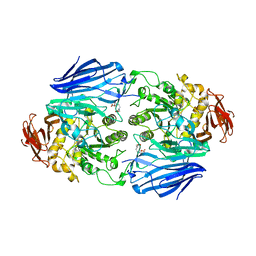 | | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase | | Descriptor: | Alpha-glucosidase, SODIUM ION, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Okuyama, M, Yoshida, T, Hondoh, H, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase
To be Published
|
|
3W37
 
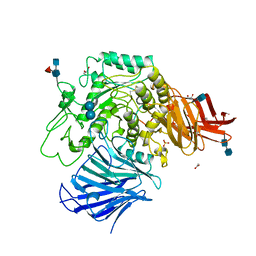 | | Sugar beet alpha-glucosidase with acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for the recognition of long-chain substrates by plant & alpha-glucosidase
J.Biol.Chem., 288, 2013
|
|
3W38
 
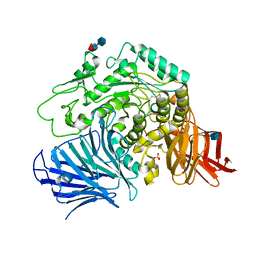 | | Sugar beet alpha-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-glucosidase, SULFATE ION, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular basis for the recognition of long-chain substrates by plant & alpha-glucosidase
J.Biol.Chem., 288, 2013
|
|
3W7W
 
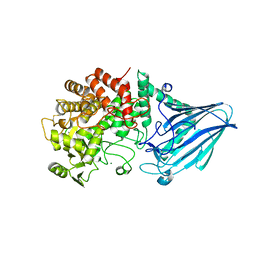 | | Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Uncharacterized protein YgjK, ... | | Authors: | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-08 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WEO
 
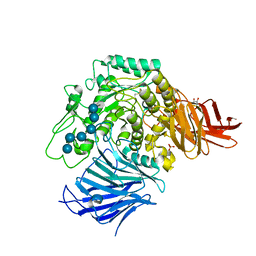 | | Sugar beet alpha-glucosidase with acarviosyl-maltohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3WEM
 
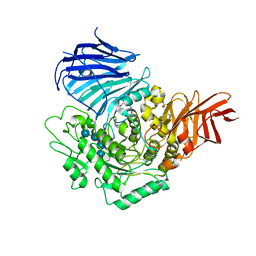 | | Sugar beet alpha-glucosidase with acarviosyl-maltotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3W7X
 
 | | Crystal structure of E. coli YgjK D324N complexed with melibiose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-08 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WEL
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltotriose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3WEN
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
