2IPC
 
 | | Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer | | Descriptor: | Preprotein translocase SecA subunit | | Authors: | Vassylyev, D.G, Mori, H, Vassylyeva, M.N, Tsukazaki, T, Kimura, Y, Tahirov, T.H, Ito, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer.
J.Mol.Biol., 364, 2006
|
|
4YLA
 
 | | Crystal structure of the indole prenyltransferase MpnD complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, Aromatic prenyltransferase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-05 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4YL7
 
 | |
4YZJ
 
 | |
4YZK
 
 | |
4YZL
 
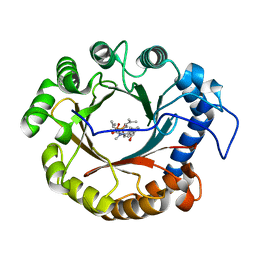 | | Crystal structure of the indole prenyltransferase TleC complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Tryptophan dimethylallyltransferase | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
3WD7
 
 | | Type III polyketide synthase | | Descriptor: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WD8
 
 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
1WRK
 
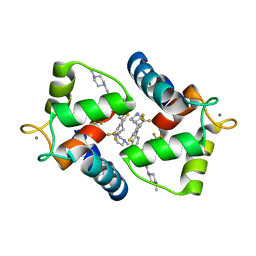 | | Crystal structure of the N-terminal domain of human cardiac troponin C in complex with trifluoperazine (orthrombic crystal form) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Troponin C, ... | | Authors: | Takeda, S, Igarashi, T, Oishi, Y, Mori, H. | | Deposit date: | 2004-10-20 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the N-terminal domain of human cardiac troponin C in complex with trifluoperazine
To be Published
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
1X03
 
 | | Crystal structure of endophilin BAR domain | | Descriptor: | SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S, Sone, M, Kamioka, Y, Mori, H, Mochizuki, N. | | Deposit date: | 2005-03-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
1WRL
 
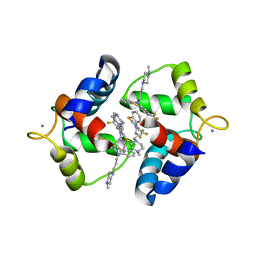 | | Crystal structure of the N-terminal domain of human cardiac troponin C in complex with trifluoperazine (monoclinic crystal form) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Troponin C, ... | | Authors: | Takeda, S, Igarashi, T, Oishi, Y, Mori, H. | | Deposit date: | 2004-10-20 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the N-terminal domain of human cardiac troponin C in complex with trifluoperazine
To be Published
|
|
2BEC
 
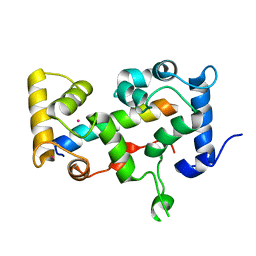 | | Crystal structure of CHP2 in complex with its binding region in NHE1 and insights into the mechanism of pH regulation | | Descriptor: | Calcineurin B homologous protein 2, Sodium/hydrogen exchanger 1, YTTRIUM (III) ION | | Authors: | Ben Ammar, Y, Takeda, S, Hisamitsu, T, Mori, H, Wakabayashi, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation
Embo J., 25, 2006
|
|
1X04
 
 | | Crystal structure of endophilin BAR domain (mutant) | | Descriptor: | SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S, Sone, M, Kamioka, Y, Mori, H, Mochizuki, N. | | Deposit date: | 2005-03-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
3WXZ
 
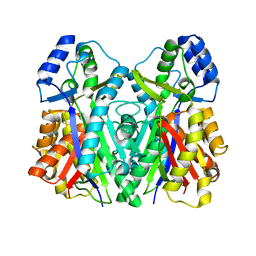 | | The structure of the I375F mutant of CsyB | | Descriptor: | Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WY0
 
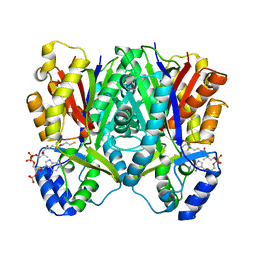 | | The I375W mutant of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WXY
 
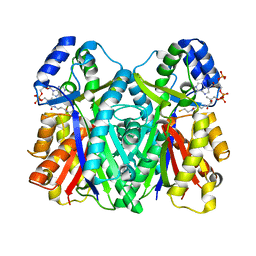 | | Crystal structure of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3X27
 
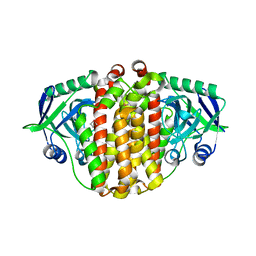 | | Structure of McbB in complex with tryptophan | | Descriptor: | Cucumopine synthase, TRYPTOPHAN | | Authors: | Mori, T, Sahashi, S, Morita, H, Abe, I. | | Deposit date: | 2014-12-10 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structural Basis for beta-Carboline Alkaloid Production by the Microbial Homodimeric Enzyme McbB
Chem.Biol., 22, 2015
|
|
8H25
 
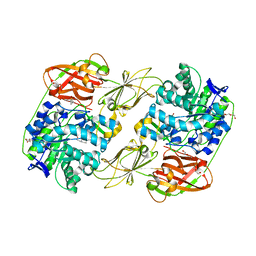 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
8IDS
 
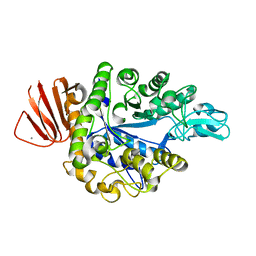 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8IBK
 
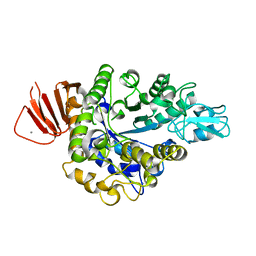 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
2ZJS
 
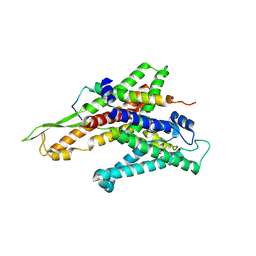 | | Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment | | Descriptor: | Fab56 (heavy chain), Fab56 (light chain), Preprotein translocase SecE subunit, ... | | Authors: | Tsukazaki, T, Mori, H, Fukai, S, Ishitani, R, Perederina, A, Vassylyev, D.G, Ito, K, Nureki, O. | | Deposit date: | 2008-03-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational transition of Sec machinery inferred from bacterial SecYE structures
Nature, 455, 2008
|
|
5AXV
 
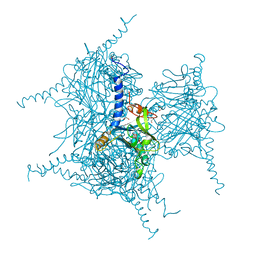 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
5AXU
 
 | | Crystal Structure of Cypovirus Polyhedra R13A Mutant | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
5YRB
 
 | | Crystal Structure of Oxidized Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
