1EID
 
 | |
1WLS
 
 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1EIC
 
 | |
1WPW
 
 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
4OOQ
 
 | | apo-dUTPase from Arabidopsis thaliana | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, SULFATE ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
1IZQ
 
 | | F46V mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IZR
 
 | | F46A mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IZP
 
 | | F46L mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1VDZ
 
 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
1FS3
 
 | |
1G2U
 
 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4OOP
 
 | | Arabidopsis thaliana dUTPase with with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
1GC8
 
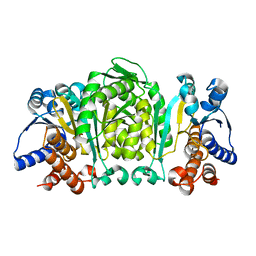 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO PHE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-27 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EIE
 
 | |
1EP9
 
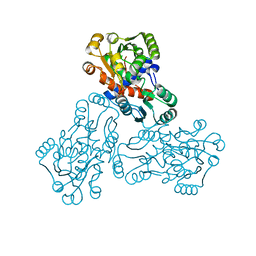 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CONFORMATIONAL CHANGE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
1XAB
 
 | |
1XAA
 
 | |
1GC9
 
 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2ZVK
 
 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1XER
 
 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
2ZVM
 
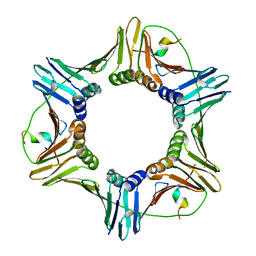 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
2ZVL
 
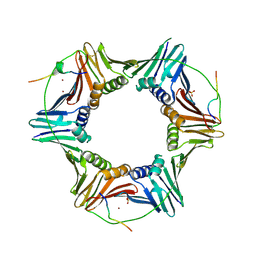 | | Crystal structure of PCNA in complex with DNA polymerase kappa fragment | | Descriptor: | DNA polymerase kappa, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
2Z47
 
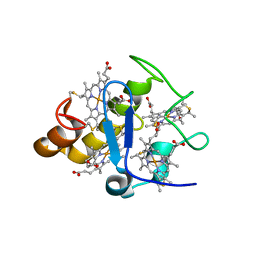 | |
1VGJ
 
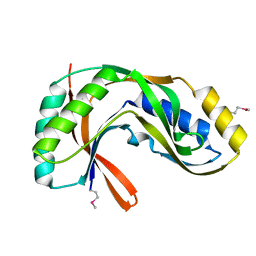 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
