6S8M
 
 | | S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Moores, C.A, von Loeffelholz, O. | | Deposit date: | 2019-07-10 | | Release date: | 2019-08-21 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM Structure (4.5- angstrom ) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance.
J.Mol.Biol., 431, 2019
|
|
5MM7
 
 | |
5MM4
 
 | |
5JCO
 
 | | Structure and dynamics of single-isoform recombinant neuronal human tubulin | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Vemu, A, Atherton, J, Spector, J.O, Szyk, A, Moores, C.A, Roll-Mecak, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and Dynamics of Single-isoform Recombinant Neuronal Human Tubulin.
J.Biol.Chem., 291, 2016
|
|
6V92
 
 | | RSC-NCP | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC14, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
8P94
 
 | |
6V8O
 
 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
2J8P
 
 | | NMR structure of C-terminal domain of human CstF-64 | | Descriptor: | CLEAVAGE STIMULATION FACTOR 64 KDA SUBUNIT | | Authors: | Qu, X, Perez-Canadillas, J.M, Agrawal, S, De Baecke, J, Cheng, H, Varani, G, Moore, C. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domains of Vertebrate Cstf-64 and its Yeast Orthologue RNA15 Form a New Structure Critical for Mrna 3'-End Processing.
J.Biol.Chem., 282, 2007
|
|
4UXR
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UY0
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXS
 
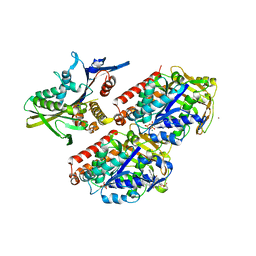 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
2OSR
 
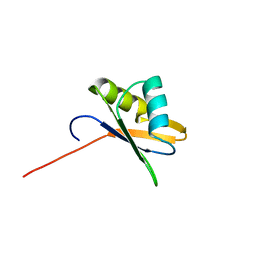 | | NMR Structure of RRM-2 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
2OSQ
 
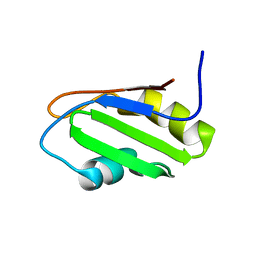 | | NMR Structure of RRM-1 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
1QAG
 
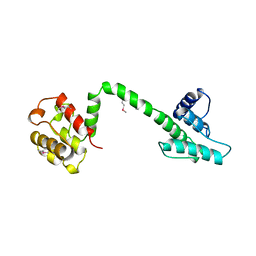 | | Actin binding region of the dystrophin homologue utrophin | | Descriptor: | UTROPHIN ACTIN BINDING REGION | | Authors: | Keep, N.H, Winder, S.J, Moores, C.A, Walke, S, Norwood, F.L.M, Kendrick-Jones, J. | | Deposit date: | 1999-03-05 | | Release date: | 2000-01-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer
Structure Fold.Des., 7, 1999
|
|
4UXP
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXO
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXY
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ALPHA TUBULIN, BETA TUBULIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
1FA0
 
 | | STRUCTURE OF YEAST POLY(A) POLYMERASE BOUND TO MANGANATE AND 3'-DATP | | Descriptor: | 3'-DEOXYADENOSINE, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Bard, J, Zhelkovsky, A.M, Helmling, S, Moore, C.L, Bohm, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP.
Science, 289, 2000
|
|
8RC1
 
 | |
5M54
 
 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6ZPI
 
 | | Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZPH
 
 | | Kinesin binding protein complexed with Kif15 motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZPG
 
 | | Kinesin binding protein (KBP) | | Descriptor: | KIF-binding protein | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6YW6
 
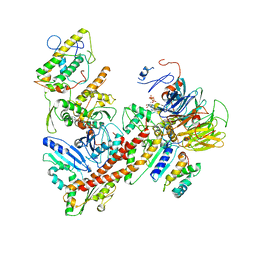 | | Cryo-EM structure of the ARP2/3 1B5CL isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARPC1B, Actin-related protein 2, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
6YW7
 
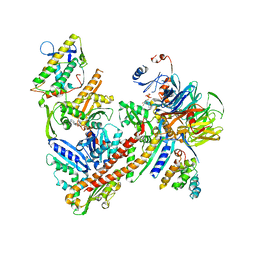 | | Cryo-EM structure of the ARP2/3 1A5C isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
