2ANX
 
 | |
254D
 
 | |
275D
 
 | |
1P36
 
 | | T4 LYOSZYME CORE REPACKING MUTANT I100V/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
3C83
 
 | |
3C81
 
 | |
3C8S
 
 | |
3C82
 
 | | Bacteriophage lysozyme T4 lysozyme mutant K85A/R96H | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C7Y
 
 | | Mutant R96A OF T4 lysozyme in wildtype background at 298K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3CDR
 
 | | R96Q Mutant of wildtype phage T4 lysozyme at 298 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-27 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3CDO
 
 | |
3CDQ
 
 | |
3C8Q
 
 | |
3CDT
 
 | |
3CDV
 
 | |
3C80
 
 | | T4 Lysozyme mutant R96Y at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8R
 
 | |
3C7W
 
 | |
3C7Z
 
 | | T4 lysozyme mutant D89A/R96H at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
1P46
 
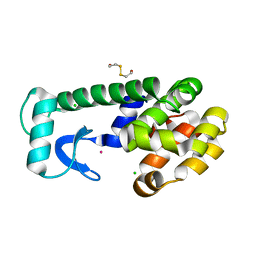 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P7S
 
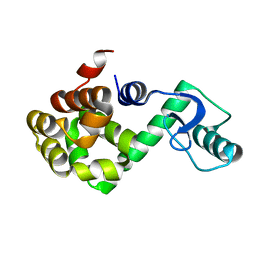 | | T4 LYSOZYME CORE REPACKING MUTANT V103I/TA | | Descriptor: | LYSOZYME | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1PQD
 
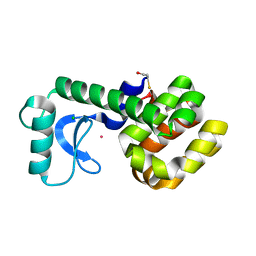 | | T4 LYSOZYME CORE REPACKING MUTANT CORE10/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1PQO
 
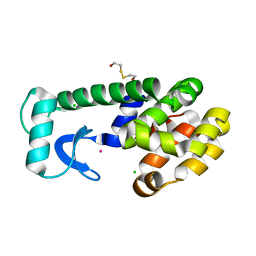 | | T4 Lysozyme Core Repacking Mutant L118I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1PQI
 
 | | T4 LYSOZYME CORE REPACKING MUTANT I118L/CORE7/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1P64
 
 | | T4 LYSOZYME CORE REPACKING MUTANT L133F/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-28 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
