1QZ9
 
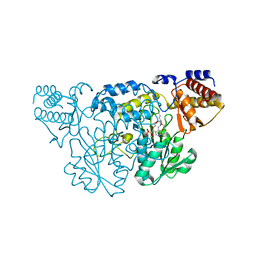 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
1AFV
 
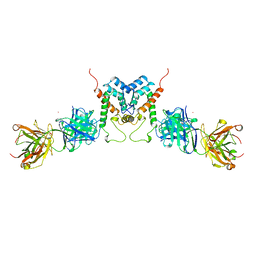 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
8SBF
 
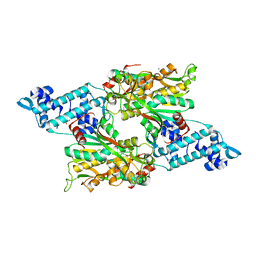 | | Full-length structure of the LysR-type transcriptional regulator, ACIAD0746, from Acinetobacter baylyi | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative transcriptional regulator (LysR family), ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E, Galloway, N.R. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
8SBH
 
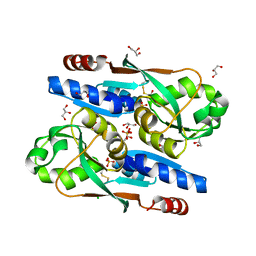 | | YeiE effector binding domain from E. coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
3FN8
 
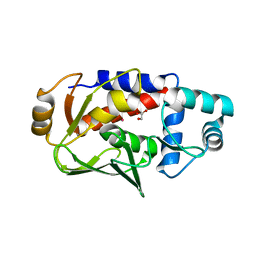 | |
3K1N
 
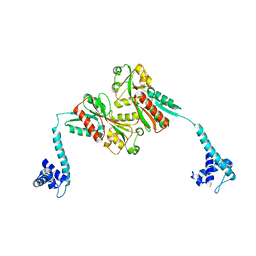 | |
4IHS
 
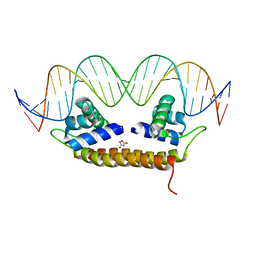 | | Crystal Structure of BenM_DBD/catB site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, MALONATE ION, SODIUM ION, ... | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IHT
 
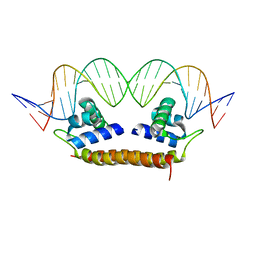 | | Crystal Structure of BenM_DBD/benA site 1 DNA Complex | | Descriptor: | HTH-type transcriptional regulator BenM, benA site 1 DNA, benA site 1 DNA - complement | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2012-12-19 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
3M1E
 
 | | Crystal Structure of BenM_DBD | | Descriptor: | HTH-type transcriptional regulator benM, SODIUM ION | | Authors: | Alanazi, A, Momany, C, Neidle, E.L. | | Deposit date: | 2010-03-04 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the unbound LysR-type transcriptional regulator BenM DNA binding domain
Acta Crystallogr.,Sect.D, 2013
|
|
1KNW
 
 | | Crystal structure of diaminopimelate decarboxylase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Diaminopimelate decarboxylase, LITHIUM ION, ... | | Authors: | Levdikov, V, Blagova, L, Bose, N, Momany, C. | | Deposit date: | 2001-12-19 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diaminopimelate Decarboxylase uses a Versatile Active Site for Stereospecific Decarboxylation
To be Published
|
|
1KO0
 
 | | Crystal Structure of a D,L-lysine complex of diaminopimelate decarboxylase | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, LYSINE, ... | | Authors: | Levdikov, V, Blagova, L, Bose, N, Momany, C. | | Deposit date: | 2001-12-19 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diaminopimelate Decarboxylase uses a Versatile Active Site for Stereospecific Decarboxylation
To be Published
|
|
3FDD
 
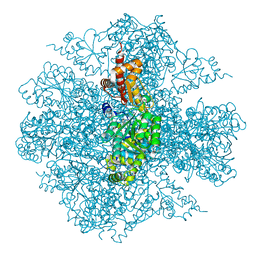 | | The Crystal Structure of the Pseudomonas dacunhae Aspartate-Beta-Decarboxylase Reveals a Novel Oligomeric Assembly for a Pyridoxal-5-Phosphate Dependent Enzyme | | Descriptor: | ACETATE ION, CHLORIDE ION, L-aspartate-beta-decarboxylase, ... | | Authors: | Lima, S, Sundararaju, B, Huang, C, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the Pseudomonas dacunhae aspartate-beta-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme.
J.Mol.Biol., 388, 2009
|
|
2F7C
 
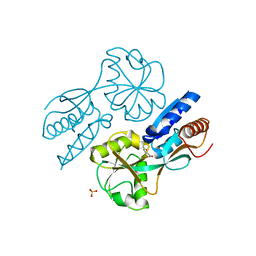 | | CatM effector binding domain with its effector cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, HTH-type transcriptional regulator catM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F78
 
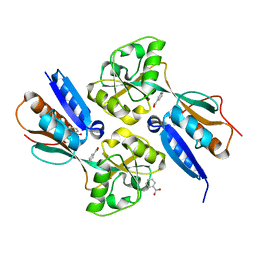 | | BenM effector binding domain with its effector benzoate | | Descriptor: | BENZOIC ACID, HTH-type transcriptional regulator benM | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F7A
 
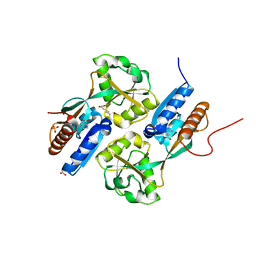 | | BenM effector binding domain with its effector, cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, ACETATE ION, BENZOIC ACID, ... | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
3K1M
 
 | | Crystal Structure of full-length BenM, R156H mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ruangprasert, A, Momany, C, Neidle, E.L, Craven, S.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Full-length BenM
To be Published
|
|
3GLB
 
 | | Crystal structure of the effector binding domain of a CATM variant (R156H) | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, GLYCEROL, HTH-type transcriptional regulator catM, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
3K1P
 
 | |
3E9K
 
 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
2F97
 
 | | Effector Binding Domain of BenM (crystals generated from high pH conditions) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, HTH-type transcriptional regulator benM, ... | | Authors: | Ezezika, O.C, Haddad, S, Neidle, E.L, Momany, C. | | Deposit date: | 2005-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligomerization of BenM, a LysR-type transcriptional regulator: structural basis for the aggregation of proteins in this family.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2F7B
 
 | | CatM effector binding domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator catM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F6G
 
 | | BenM effector binding domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-29 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2F8D
 
 | | BenM effector-Binding domain crystallized from high pH conditions | | Descriptor: | BENZOIC ACID, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ezezika, O.C, Haddad, S, Neidle, E.L, Momany, C. | | Deposit date: | 2005-12-02 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oligomerization of BenM, a LysR-type transcriptional regulator: structural basis for the aggregation of proteins in this family.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2H98
 
 | |
