1IRE
 
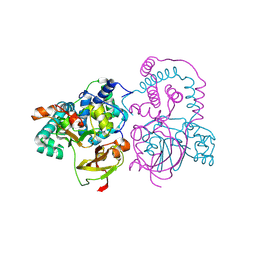 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | COBALT (II) ION, Nitrile Hydratase | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | Deposit date: | 2001-10-01 | | Release date: | 2002-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
8K4R
 
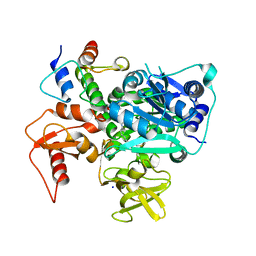 | | Structure of VinM-VinL complex | | Descriptor: | Acyl-carrier-protein, Non-ribosomal peptide synthetase, SODIUM ION, ... | | Authors: | Miyanaga, A, Nagata, K, Nakajima, J, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2023-07-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
6M01
 
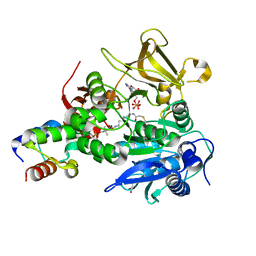 | | The structure of HitB-HitD complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Miyanaga, A, Kurihara, S, Kudo, F, Eguchi, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Complex of Adenylation Domain and Carrier Protein by Using Pantetheine Cross-Linking Probe.
Acs Chem.Biol., 15, 2020
|
|
8K5S
 
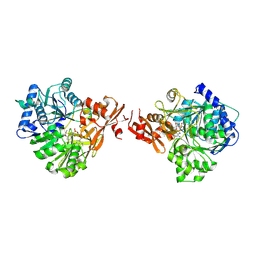 | | The structure of EntE with 3-(prop-2-yn-1-yloxy)benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-prop-2-ynoxyphenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
8K5T
 
 | | The structure of EntE with2-methyl-3-chloro-benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-chloranyl-2-methyl-phenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
1WD3
 
 | | Crystal structure of arabinofuranosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
1WD4
 
 | | Crystal structure of arabinofuranosidase complexed with arabinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
2D43
 
 | | Crystal structure of arabinofuranosidase complexed with arabinotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
2D44
 
 | | Crystal structure of arabinofuranosidase complexed with arabinofuranosyl-alpha-1,2-xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-2)-alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
1UGS
 
 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGR
 
 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGQ
 
 | | Crystal structure of apoenzyme of Co-type nitrile hydratase | | Descriptor: | Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
5CZD
 
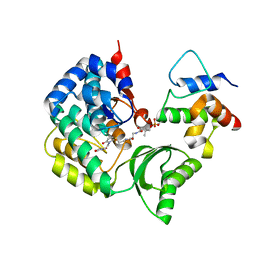 | | The complex structure of VinK with VinL | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, Acyl-carrier-protein, ... | | Authors: | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1UGP
 
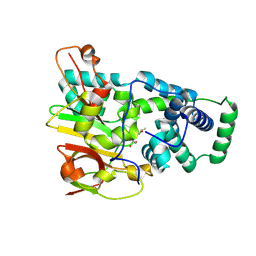 | | Crystal structure of Co-type nitrile hydratase complexed with n-butyric acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
7F2R
 
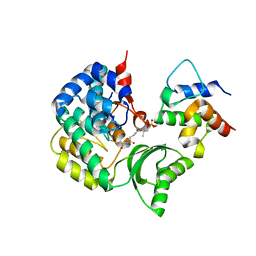 | | Crystal structure of VinK-VinL covalent complex formed with a pantetheineamide cross-linking probe | | Descriptor: | Acyl-carrier-protein, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Miyanaga, A, Ouchi, R, Kudo, F, Eguchi, T. | | Deposit date: | 2021-06-14 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex structure of the acyltransferase VinK and the carrier protein VinL with a pantetheine cross-linking probe.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5CZC
 
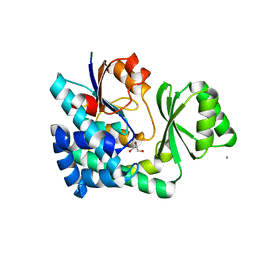 | | The structure of VinK | | Descriptor: | CALCIUM ION, GLYCEROL, Malonyl-CoA-[acyl-carrier-protein] transacylase | | Authors: | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6K96
 
 | | Crystal structure of Ari2 | | Descriptor: | Five-membered-cyclitol-phosphate synthase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Miyanaga, A, Tsunoda, T, Kudo, F, Eguchi, T. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereochemistry in the Reaction of themyo-Inositol Phosphate Synthase Ortholog Ari2 during Aristeromycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6IYL
 
 | | The structure of EntE with 3-cyanobenzoyl adenylate analog | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(3-cyanobenzene-1-carbonyl)sulfamoyl]adenosine | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6IYK
 
 | | The structure of EntE with 2-nitrobenzoyl adenylate analog | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex, 5'-O-[(2-nitrobenzene-1-carbonyl)sulfamoyl]adenosine | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An Engineered Aryl Acid Adenylation Domain with an Enlarged Substrate Binding Pocket.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3WFI
 
 | | The crystal structure of D-mandelate dehydrogenase | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
5Y1I
 
 | | The crystal structure of GfsF | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2017-07-20 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition by a Dual-Function P450 Monooxygenase GfsF Involved in FD-891 Biosynthesis
Chembiochem, 18, 2017
|
|
3WFJ
 
 | | The complex structure of D-mandelate dehydrogenase with NADH | | Descriptor: | 2-dehydropantoate 2-reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
5ZK4
 
 | | The structure of DSZS acyltransferase with carrier protein | | Descriptor: | DisA protein, DisD protein, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Miyanaga, A, Ouchi, R, Kudo, F, Eguchi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of protein-protein interactions between a trans-acting acyltransferase and acyl carrier protein in polyketide disorazole biosynthesis
J. Am. Chem. Soc., 140, 2018
|
|
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WV4
 
 | | Crystal structure of VinN | | Descriptor: | Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
