4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
5MFD
 
 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
4DB8
 
 | | Designed Armadillo-repeat Protein | | Descriptor: | Armadillo-repeat Protein, MAGNESIUM ION | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
2CJX
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-09 | | Release date: | 2006-06-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
5LW1
 
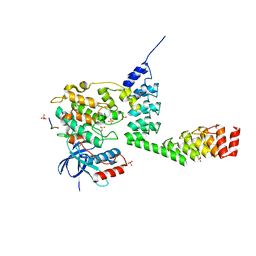 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
2CJY
 
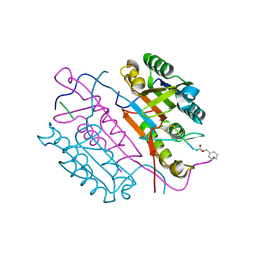 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-09 | | Release date: | 2006-06-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
4DB6
 
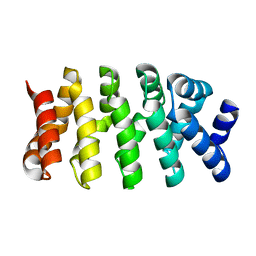 | | Designed Armadillo repeat protein (YIIIM3AII) | | Descriptor: | Armadillo repeat protein | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB9
 
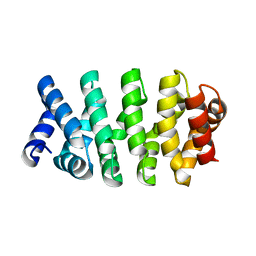 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
5MDI
 
 | | Crystal structure of TDP-43 N-terminal domain at 2.1 A resolution | | Descriptor: | ACETATE ION, TAR DNA-binding protein 43 | | Authors: | Afroz, T, Hock, E.-M, Ernst, P, Foglieni, C, Jambeau, M, Gilhespy, L, Laferriere, F, Maniecka, Z, Plueckthun, A, Mittl, P, Paganetti, P, Allain, F.H.T, Polymenidou, M. | | Deposit date: | 2016-11-11 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation.
Nat Commun, 8, 2017
|
|
2Y0B
 
 | | Caspase-3 in Complex with an Inhibitory DARPin-3.4_S76R | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CASPASE-3, ... | | Authors: | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | Deposit date: | 2010-12-01 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific Inhibition of Caspase-3 by a Competitive Darpin: Molecular Mimicry between Native and Designed Inhibitors.
Structure, 21, 2013
|
|
2XZD
 
 | | Caspase-3 in Complex with an Inhibitory DARPin-3.4 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CASPASE-3, ... | | Authors: | Barandun, J, Schroeder, T, Mittl, P, Grutter, M.G. | | Deposit date: | 2010-11-24 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific Inhibition of Caspase-3 by a Competitive Darpin: Molecular Mimicry between Native and Designed Inhibitors.
Structure, 21, 2013
|
|
5MFC
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4CG4
 
 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
2XEE
 
 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C DARPIN MUTANT5 | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2XEH
 
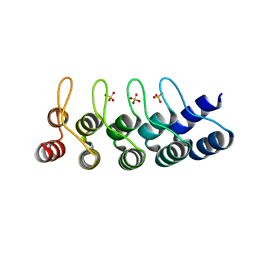 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C MUT6, SULFATE ION | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
5OD9
 
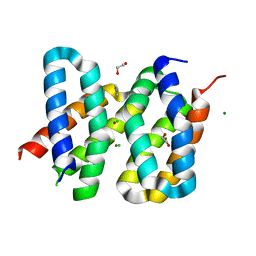 | | Structure of the engineered metalloesterase MID1sc9 | | Descriptor: | (2~{S})-2-phenylpropanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Studer, S, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2017-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4BS0
 
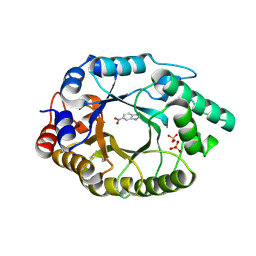 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
2XYH
 
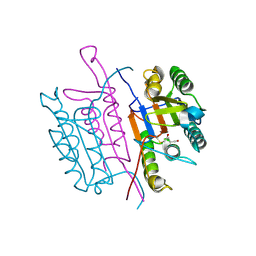 | | Caspase-3:CAS60254719 | | Descriptor: | 5-CHLORO-4-OXOPENTANOIC ACID, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-17 | | Release date: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XYP
 
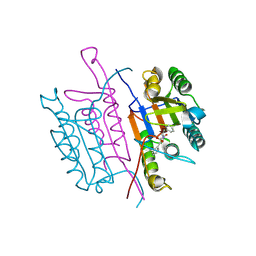 | | Caspase-3:CAS26049945 | | Descriptor: | CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17, PHENYLMETHYL N-[(2S)-4-CHLORO-3-OXO-1-PHENYL-BUTAN-2-YL]CARBAMATE | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-18 | | Release date: | 2011-08-17 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XYG
 
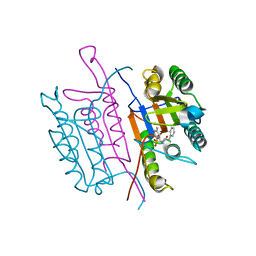 | | Caspase-3:CAS329306 | | Descriptor: | CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-17 | | Release date: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XEN
 
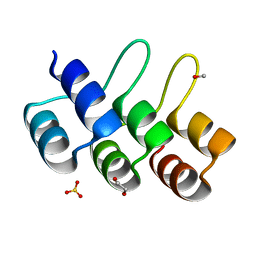 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, NI1C MUT4, ... | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
3ZEV
 
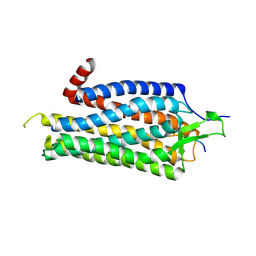 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BUO
 
 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
