4JRR
 
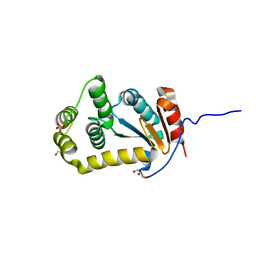 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
4JXQ
 
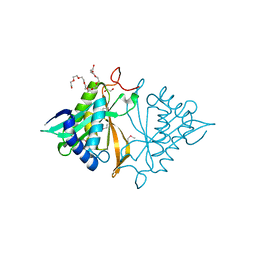 | | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti 1021 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CITRATE ANION, ... | | Authors: | Majorek, K.A, Cooper, D.R, Osinski, T, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti 1021
To be Published
|
|
2FQ4
 
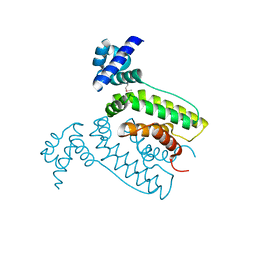 | | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Zhang, R, Wu, R, Moy, S, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus
To be Published
|
|
4JWV
 
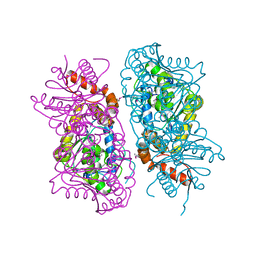 | | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Short chain enoyl-CoA hydratase | | Authors: | Cooper, D.R, Mikolajczak, K, Cymborowski, M, Grabowski, M, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JXR
 
 | | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CITRATE ANION, ... | | Authors: | Majorek, K.A, Cooper, D.R, Porebski, P.J, Konina, K, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA
To be Published
|
|
2FU2
 
 | | Crystal structure of protein SPy2152 from Streptococcus pyogenes | | Descriptor: | Hypothetical protein SPy2152 | | Authors: | Chang, C, Cymborowski, M, Otwinowski, Z, Minor, W, Lezondra, L.-E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-25 | | Release date: | 2006-03-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of pyogenecin immunity protein, a novel bacteriocin-like immunity protein from Streptococcus pyogenes.
Bmc Struct.Biol., 9, 2009
|
|
4JHX
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoylphosphate and arginine | | Descriptor: | ARGININE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Bacal, P, Winsor, J, Grimshaw, S, Grabowski, M, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4JXU
 
 | | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP | | Descriptor: | Putative aminotransferase | | Authors: | Cooper, D.R, Cymborowski, M.T, Majorek, K.A, Niedzialkowska, E, Porebski, P.J, Stead, M, Hammonds, J, Seidel, R, Ahmed, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP
To be Published
|
|
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2O6T
 
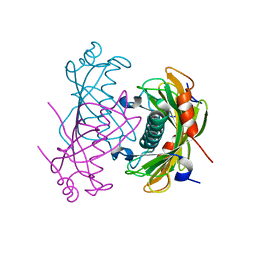 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (P2221). | | Descriptor: | CHLORIDE ION, THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3OWC
 
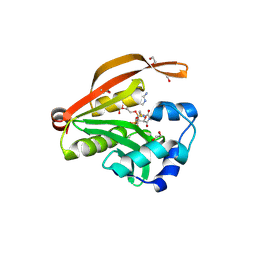 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
4KUA
 
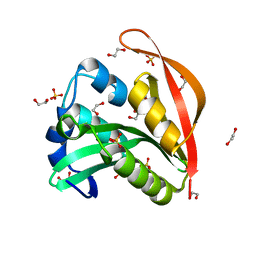 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 | | Descriptor: | 1,2-ETHANEDIOL, GNAT superfamily acetyltransferase PA4794, SULFATE ION | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
2IAI
 
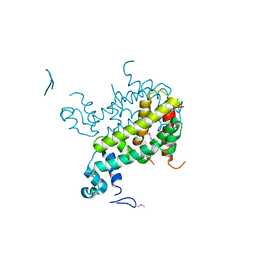 | | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3 | | Descriptor: | Putative transcriptional regulator SCO3833 | | Authors: | Zimmerman, M.D, Xu, X, Wang, S, Gu, J, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3
To be Published
|
|
2O5U
 
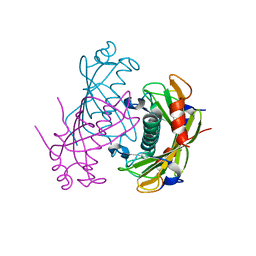 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (C222). | | Descriptor: | Thioesterase | | Authors: | Chruszcz, M, Wang, S, Evdokimova, E, Koclega, K.D, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O8N
 
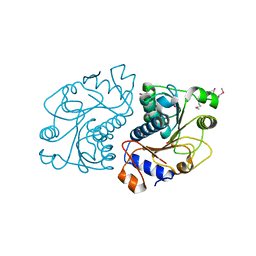 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein | | Descriptor: | ApoA-I binding protein, CHLORIDE ION, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Zheng, H, Chruszcz, M, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Apolipoprotein A-I Binding Protein, a Novel Phosphoprotein with a Potential Role in Sperm Capacitation.
Endocrinology, 149, 2008
|
|
2I8E
 
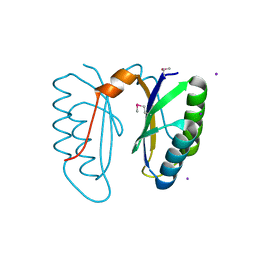 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
4JYJ
 
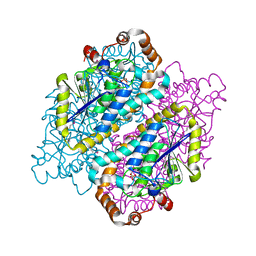 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JYK
 
 | | Structure of E. coli Transcriptional Regulator RutR with bound uracil | | Descriptor: | HTH-type transcriptional regulator RutR, SULFATE ION, URACIL | | Authors: | Cooper, D.R, Knapik, A.A, Petkowski, J.J, Porebski, P.J, Osinski, T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of E. coli Transcriptional Regulator RutR with bound uracil
To be Published
|
|
2O0Y
 
 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | Descriptor: | Transcriptional regulator | | Authors: | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
2G7U
 
 | | 2.3 A structure of putative catechol degradative operon regulator from Rhodococcus sp. RHA1 | | Descriptor: | transcriptional regulator | | Authors: | Zheng, H, Skarina, T, Chruszcz, M, Cymborowski, M, Grabowski, M, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 A structure of putative catechol degradative operon regulator from
Rhodococcus sp. RHA1
To be Published
|
|
3QTB
 
 | | Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, Uncharacterized protein | | Authors: | Tkaczuk, K.L, Shumilin, I.A, Chruszcz, M, Cymborowski, M, Xu, X, Di Leo, R, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3QXS
 
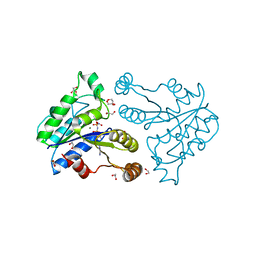 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
2O6B
 
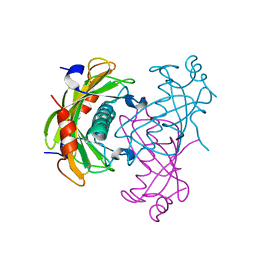 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6U
 
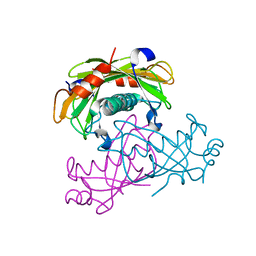 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3QXC
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
