8JUP
 
 | | Crystal structure of a receptor like kinase from rice | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LRR receptor-like serine/threonine-protein kinase FLS2, MAGNESIUM ION | | Authors: | Ming, Z, Zhao, Q. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An Active State Formation Mechanism of Receptor Kinase in Plant
Plant Commun., 2024
|
|
8JUV
 
 | | Crystal structure of a receptor like kinase with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, LRR receptor-like serine/threonine-protein kinase FLS2, MAGNESIUM ION | | Authors: | Ming, Z, Zhao, Q. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | An active State Formation Mechanism of Receptor Kinase in Plant
Plant Commun., 2024
|
|
6K62
 
 | | Crystal structure of Xanthomonas PcrK | | Descriptor: | Histidine kinase | | Authors: | Ming, Z.H, Tang, J.L, Wu, L.J, Chen, P. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the phytopathogenic bacterial sensor PcrK reveals different cytokinin recognition mechanism from the plant sensor AHK4.
J.Struct.Biol., 208, 2019
|
|
5GO3
 
 | | Crystal structure of a di-nucleotide cyclase Vibrio mutant | | Descriptor: | Cyclic GMP-AMP synthase | | Authors: | Ming, Z.H, Wang, W, Xie, Y.C, Chen, Y.C, Yan, L.M, Lou, Z.Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a di-nucleotide cyclase Vibrio mutant
To Be Published
|
|
7F52
 
 | |
9IY0
 
 | | anti-HEV mAb 8H3 | | Descriptor: | Heavy chain of 8H3 Fab, Light chain of 8H3 Fab, MAGNESIUM ION | | Authors: | Minghua, Z, Lizhi, Z, Yang, H, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
9IY2
 
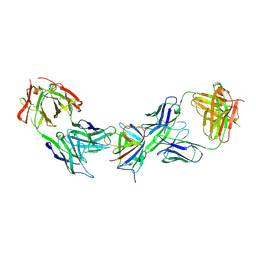 | | Immune complex of HEV-E2s, nAb 8C11 and nAb 8H3 | | Descriptor: | Heavy Chain of mAb 8C11, Heavy Chain of mAb 8H3, Light Chain of mAb 8C11, ... | | Authors: | Minghua, Z, Lizhi, Z, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.476 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
4R3D
 
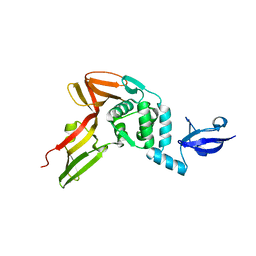 | | Crystal structure of MERS Coronavirus papain like protease | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Kong, L.Y, Wang, Q, Ming, Z.H, Shaw, N, Sun, Y.N, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Crystal Structures of Middle East Respiratory Syndrome Coronavirus Papain-like Protease and Its Complex with an Inhibitor
To be Published
|
|
7BRC
 
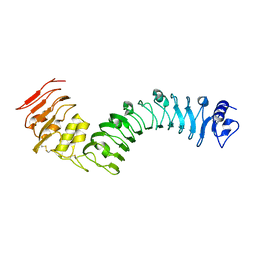 | | Crystal structure of the TMK3 LRR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-like kinase TMK3 | | Authors: | Chen, H, Kong, Y.Q, Chen, J, Li, L, Li, X.S, Yu, F, Ming, Z.H. | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the extracellular domain of the receptor-like kinase TMK3 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7XDX
 
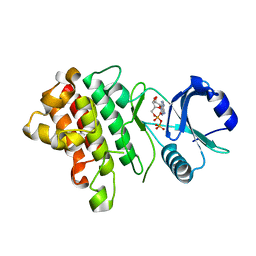 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDV
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDW
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDY
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7DK8
 
 | | Crystal structure of OsGH3-8 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable indole-3-acetic acid-amido synthetase GH3.8 | | Authors: | Zhang, Y.K, Xu, G.L, Ming, Z.H. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7YQR
 
 | | Crystal Structure of Xcc NAMPT and its complex with NAM | | Descriptor: | NICOTINAMIDE, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7YQQ
 
 | | Crystal Structure of Xcc NAMPT and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, PHOSPHATE ION, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7YQO
 
 | | Xcc Nicotinamide Phosphoribosyltransferase | | Descriptor: | GLYCEROL, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7F5W
 
 | |
8IGZ
 
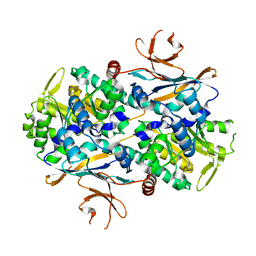 | | Xcc NAMPT Quadruple mutant | | Descriptor: | Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2023-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
6LP3
 
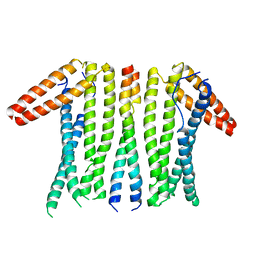 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
7YQP
 
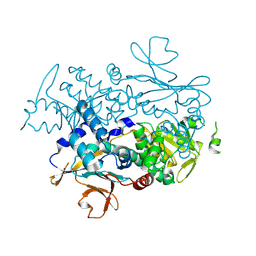 | | Xcc Nicotinamide Phosphoribosyltransferase | | Descriptor: | Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7W3R
 
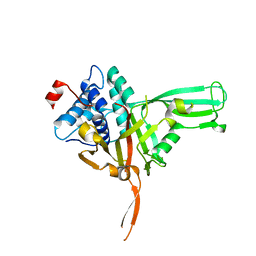 | | USP34 catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
8GYZ
 
 | |
8HOA
 
 | | ScRIPK mutant K124R | | Descriptor: | (2R,5R)-hexane-2,5-diol, ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, ... | | Authors: | Fang, J.L, Zhang, M.Q, Ming, Z.H. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
