6MUQ
 
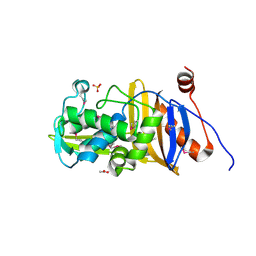 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
6W4H
 
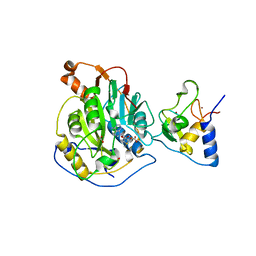 | | 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, ACETATE ION, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6W75
 
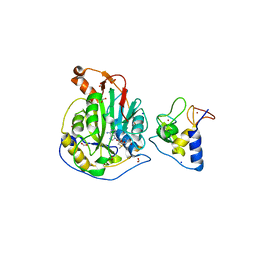 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
1G2J
 
 | | RNA OCTAMER R(CCCP*GGGG) CONTAINING PHENYL RIBONUCLEOTIDE | | Descriptor: | 5'-R(*CP*CP*CP*(PYY)P*GP*GP*GP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Matulic-Adamic, J, Wilds, C.J, Haeberli, P, Usman, N, Beigelman, L, Egli, M. | | Deposit date: | 2000-10-20 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of an RNA duplex containing phenyl-ribonucleotides, hydrophobic isosteres of the natural pyrimidines.
RNA, 6, 2000
|
|
6N7M
 
 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | Descriptor: | Hypothetical Protein CD630_05490 | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
3DZC
 
 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
3G25
 
 | | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHATE ION, ... | | Authors: | Minasov, G, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol.
TO BE PUBLISHED
|
|
4RGT
 
 | | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine.
TO BE PUBLISHED
|
|
4RH6
 
 | | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Exotoxin 3, putative | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Putative Exotoxin 3 from Staphylococcus aureus.
TO BE PUBLISHED
|
|
2GX8
 
 | | The Crystal Structure of Bacillus cereus protein related to NIF3 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NIF3-related protein, ... | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-08 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution crystal structure of Bacillus cereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain
Protein Sci., 16, 2007
|
|
2FOR
 
 | | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate | | Descriptor: | Geranyltranstransferase, ISOPENTYL PYROPHOSPHATE, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Shigella flexneri Farnesyl Pyrophosphate Synthase Complex with an Isopentenyl Pyrophosphate
To be Published
|
|
2GJV
 
 | | Crystal Structure of a Protein of Unknown Function from Salmonella typhimurium | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative cytoplasmic protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-31 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of a Hypothetical Protein from Salmonella typhimurium
To be Published
|
|
4QVR
 
 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | Descriptor: | Uncharacterized hypothetical protein FTT_1539c | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
4QWO
 
 | | 1.52 Angstrom Crystal Structure of A42R Profilin-like Protein from Monkeypox Virus Zaire-96-I-16 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-06 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the Monkeypox virus profilin-like protein A42R reveals potential functional differences from cellular profilins.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6MUK
 
 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
2I0Z
 
 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
4RO3
 
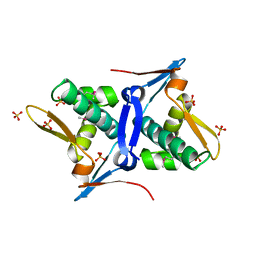 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | Hypothetical Protein, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
478D
 
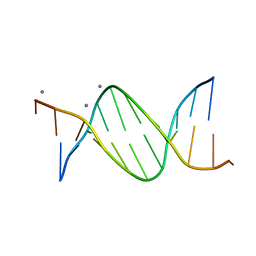 | |
4S24
 
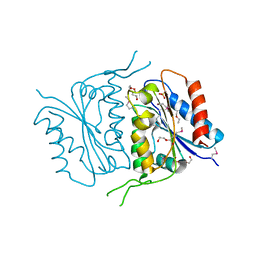 | | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Modulator of drug activity B, PENTAETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
4RWR
 
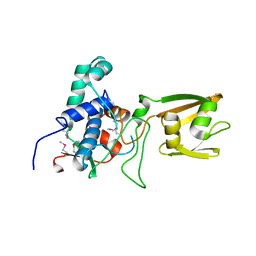 | | 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis | | Descriptor: | Stage II sporulation protein D | | Authors: | Minasov, G, Wawrzak, Z, Nocadello, S, Shuvalova, L, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
4S12
 
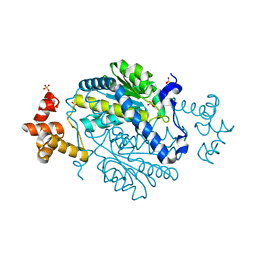 | | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetylmuramic acid 6-phosphate etherase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of N-acetylmuramic acid 6-phosphate Etherase from Yersinia enterocolitica.
TO BE PUBLISHED
|
|
476D
 
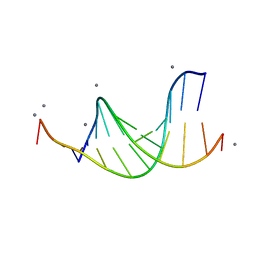 | | CALCIUM FORM OF B-DNA UNDECAMER GCGAATTCGCG | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Tereshko, V, Egli, M. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing.
J.Mol.Biol., 291, 1999
|
|
3FGX
 
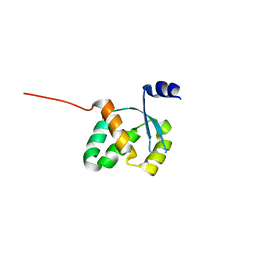 | | Structure of uncharacterised protein rbstp2171 from bacillus stearothermophilus | | Descriptor: | RBSTP2171 | | Authors: | Minasov, G, Filippova, E.V, Shuvalova, L, Moy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Uncharacterised Protein Rbstp2171 from Bacillus Stearothermophilus
To be Published
|
|
477D
 
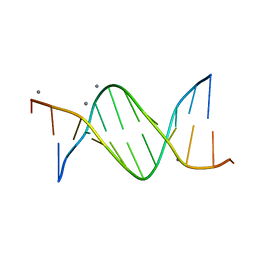 | | CALCIUM FORM OF THE B-DNA DODECAMER GGCGAATTCGCG | | Descriptor: | 5'-D(*GP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Tereshko, V, Egli, M. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing.
J.Mol.Biol., 291, 1999
|
|
7JZ0
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
