6VPU
 
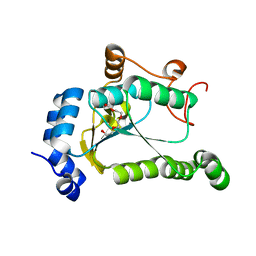 | | 1.90 Angstrom Resolution Crystal Structure Phosphoadenosine Phosphosulfate Reductase (CysH) from Vibrio vulnificus | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Phosphoadenosine phosphosulfate reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure Phosphoadenosine Phosphosulfate Reductase (CysH) from Vibrio vulnificus
To Be Published
|
|
4RS2
 
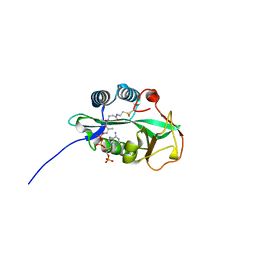 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | Descriptor: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | Authors: | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
6VBF
 
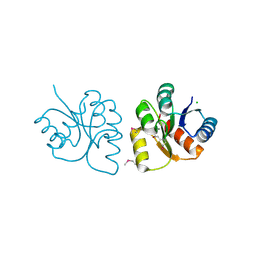 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
6VJ6
 
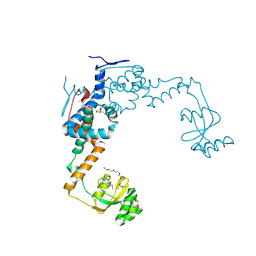 | | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, GLYCEROL, Peptidylprolyl isomerase (PrsA) | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus cereus
To Be Published
|
|
6WJI
 
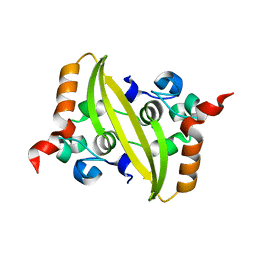 | | 2.05 Angstrom Resolution Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, Nucleoprotein | | Authors: | Minasov, G, Shuvalova, L, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Serodominant SARS-CoV-2 Nucleocapsid Peptides Map to Unstructured Protein Regions.
Microbiol Spectr, 2023
|
|
6WN8
 
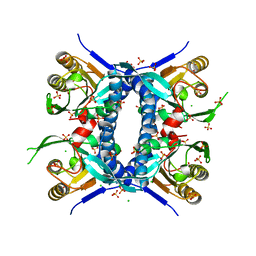 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X2D
 
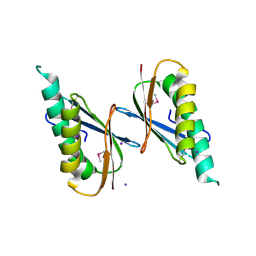 | | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae | | Descriptor: | IODIDE ION, Ribonuclease E | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-20 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae.
To Be Published
|
|
6VBB
 
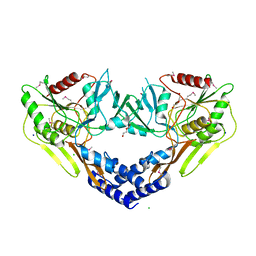 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6VJ2
 
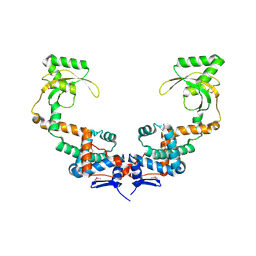 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | Descriptor: | Foldase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
6VJ4
 
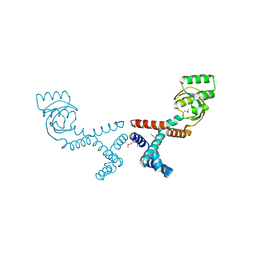 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6VPW
 
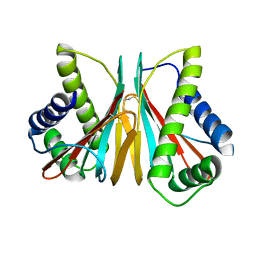 | | 1.90 Angstrom Resolution Crystal Structure Chemotaxis protein CheX from Vibrio vulnificus | | Descriptor: | Chemotaxis protein CheX | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure Chemotaxis protein CheX from Vibrio vulnificus
To Be Published
|
|
6WN5
 
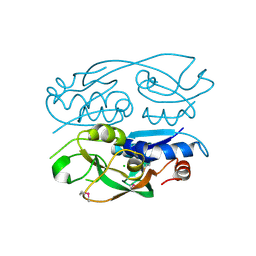 | | 1.52 Angstrom Resolution Crystal Structure of Transcriptional Regulator HdfR from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Transcriptional regulator HdfR | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6UE2
 
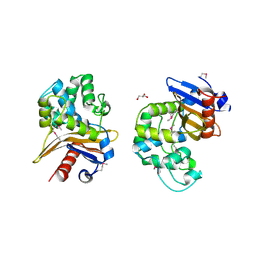 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6UOF
 
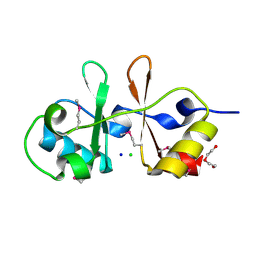 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
7KPO
 
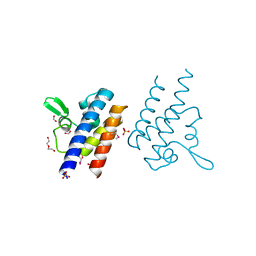 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
7L6J
 
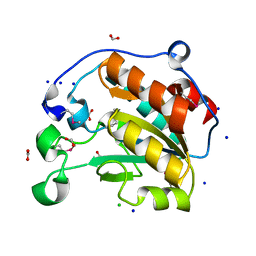 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
6WJT
 
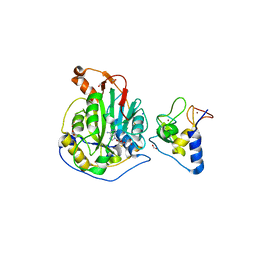 | | 2.0 Angstrom Resolution Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with S-Adenosyl-L-Homocysteine | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WY4
 
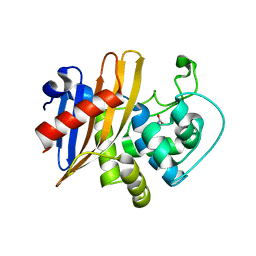 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6WKQ
 
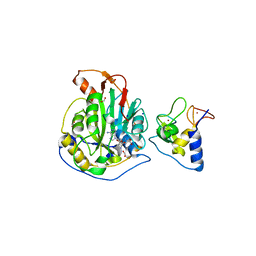 | | 1.98 Angstrom Resolution Crystal Structure of NSP16-NSP10 Heterodimer from SARS-CoV-2 in Complex with Sinefungin | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WVN
 
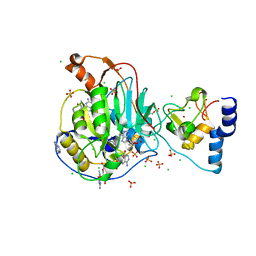 | | Crystal Structure of Nsp16-Nsp10 from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-Adenosylmethionine. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
5BXI
 
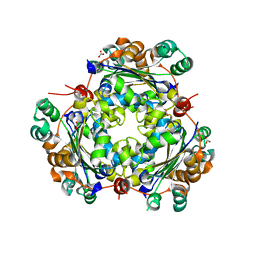 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
7K1U
 
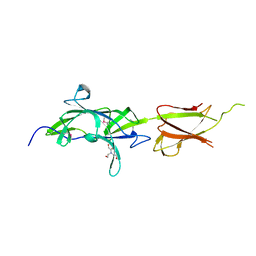 | | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Collagen-binding Adhesin | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630
To Be Published
|
|
5EQV
 
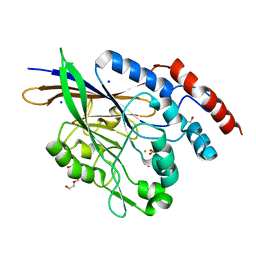 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
5DZS
 
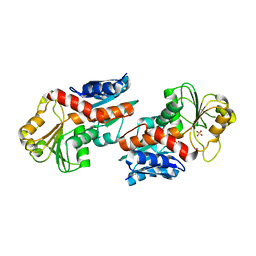 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
