4MBR
 
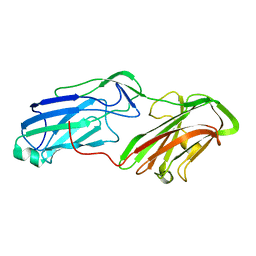 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
3GE1
 
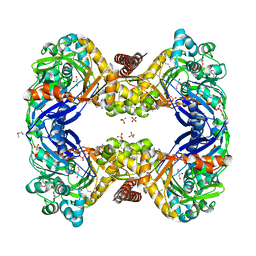 | | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Brunzelle, J, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol
To be Published
|
|
4IIW
 
 | | 2.6 Angstrom Crystal Structure of Putative yceG-like Protein lmo1499 from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Lmo1499 protein, SULFATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of Putative yceG-like Protein lmo1499 from Listeria monocytogenes
TO BE PUBLISHED
|
|
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
4MBO
 
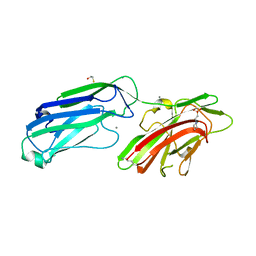 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
3TNL
 
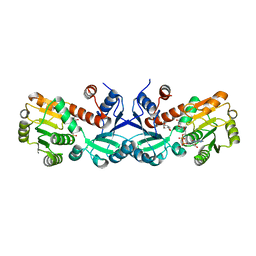 | | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD. | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.
TO BE PUBLISHED
|
|
4MGE
 
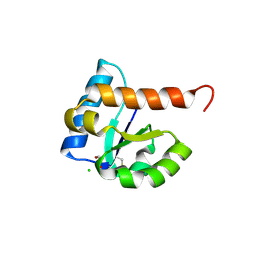 | | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PTS system, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis.
TO BE PUBLISHED
|
|
5VFB
 
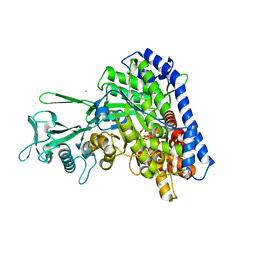 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
5VH6
 
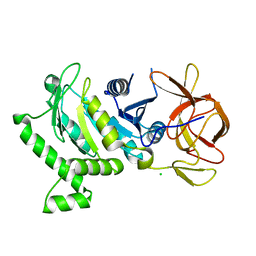 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
3N3W
 
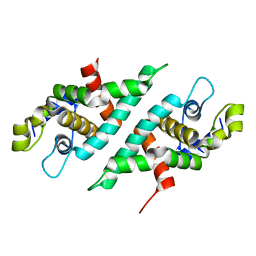 | | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni
TO BE PUBLISHED
|
|
3QM3
 
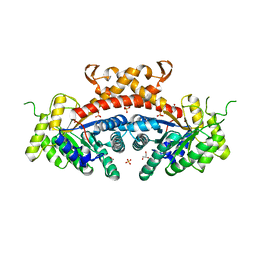 | | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fructose-bisphosphate aldolase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni
TO BE PUBLISHED
|
|
3L2I
 
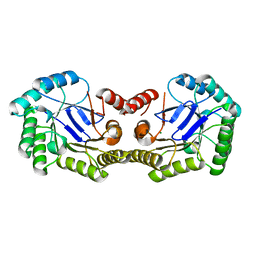 | | 1.85 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2. | | Descriptor: | 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.
Biochemistry, 50, 2011
|
|
3QFK
 
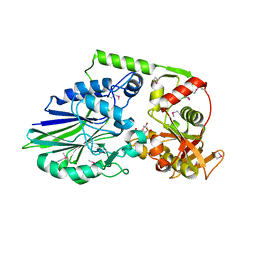 | | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Krishna, S.N, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate.
TO BE PUBLISHED
|
|
3QFH
 
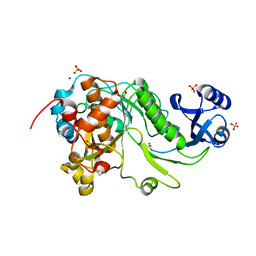 | | 2.05 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) from Staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, Epidermin leader peptide processing serine protease EpiP, GLYCEROL, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) from Staphylococcus aureus.
TO BE PUBLISHED
|
|
5W6L
 
 | | Crystal Structure of RRSP, a MARTX Toxin Effector Domain from Vibrio vulnificus CMCP6 | | Descriptor: | CHLORIDE ION, GLYCEROL, RTX repeat-containing cytotoxin, ... | | Authors: | Minasov, G, Wawrzak, Z, Biancucci, M, Shuvalova, L, Dubrovska, I, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling.
Sci Signal, 11, 2018
|
|
5TLS
 
 | | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD | | Descriptor: | (2S)-4-(6-amino-9H-purin-9-yl)-2-hydroxybutanoic acid, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD
To Be Published
|
|
3OKF
 
 | | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae.
TO BE PUBLISHED
|
|
5U2G
 
 | | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae.
To Be Published
|
|
6B8W
 
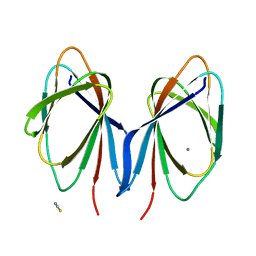 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
3PNU
 
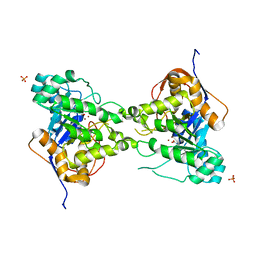 | | 2.4 Angstrom Crystal Structure of Dihydroorotase (pyrC) from Campylobacter jejuni. | | Descriptor: | Dihydroorotase, PHOSPHATE ION, ZINC ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Dihydroorotase (pyrC) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
6D0G
 
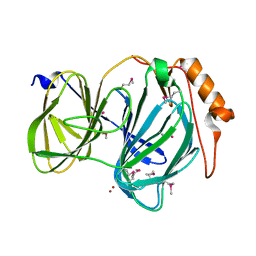 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
3ROH
 
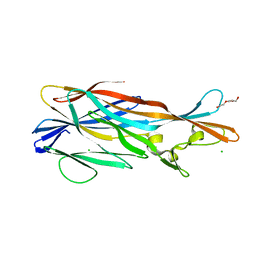 | | Crystal Structure of Leukotoxin (LukE) from Staphylococcus aureus subsp. aureus COL. | | Descriptor: | CHLORIDE ION, Leucotoxin LukEv, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3UPD
 
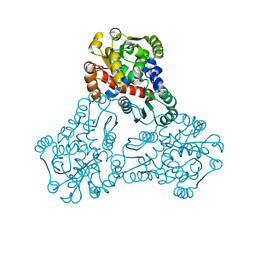 | | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus.
TO BE PUBLISHED
|
|
5T8K
 
 | | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD.
To Be Published
|
|
5TJ9
 
 | | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD | | Descriptor: | (1R,2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD
To Be Published
|
|
