1HXS
 
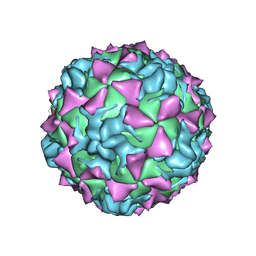 | | CRYSTAL STRUCTURE OF MAHONEY STRAIN OF POLIOVIRUS AT 2.2A RESOLUTION | | Descriptor: | GENOME POLYPROTEIN, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | Authors: | Miller, S.T, Hogle, J.M, Filman, D.J. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
8G6Z
 
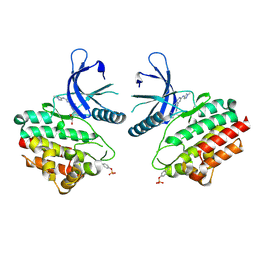 | | JAK2 crystal structure in complex with Compound 13 | | Descriptor: | (3R)-3-cyclopentyl-3-[(4M)-4-{5-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
8G8O
 
 | | The crystal structure of JAK2 in complex with Compound 31 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Tyrosine-protein kinase JAK2, ... | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
8G8X
 
 | | X-ray co-crystal structure of compound 27 in with complex JAK2 | | Descriptor: | 3-cyclopropyl-1-{5-methyl-2-[(3-methyl-1,2-thiazol-5-yl)amino]pyrimidin-4-yl}azetidin-3-ol, Tyrosine-protein kinase JAK2 | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
3QMQ
 
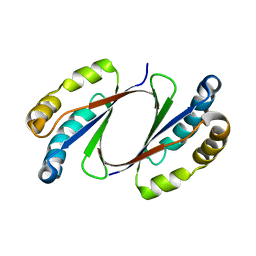 | | Crystal Structure of E. coli LsrG | | Descriptor: | Autoinducer-2 (AI-2) modifying protein LsrG | | Authors: | Miller, S.T, Russell, C. | | Deposit date: | 2011-02-04 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processing the Interspecies Quorum-sensing Signal Autoinducer-2 (AI-2): CHARACTERIZATION OF PHOSPHO-(S)-4,5-DIHYDROXY-2,3-PENTANEDIONE ISOMERIZATION BY LsrG PROTEIN.
J.Biol.Chem., 286, 2011
|
|
4P2V
 
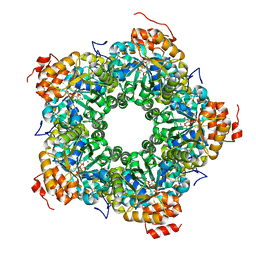 | | Structure of the AI-2 processing enzyme LsrF in complex with the product of the LsrG reaction P-HPD | | Descriptor: | (3R)-3-hydroxy-2,4-dioxopentyl dihydrogen phosphate, Uncharacterized aldolase LsrF | | Authors: | Miller, S.T, Oh, I.K, Xavier, K.B. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3EJW
 
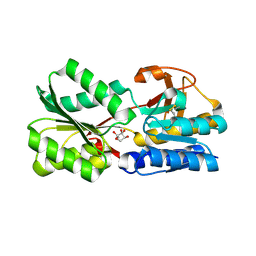 | | Crystal Structure of the Sinorhizobium meliloti AI-2 receptor, SmLsrB | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, SmLsrB | | Authors: | Miller, S.T, McAuley, J.R, Pereira, C, Xavier, K.B, Taga, M.E. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria.
Mol.Microbiol., 70, 2008
|
|
1TJY
 
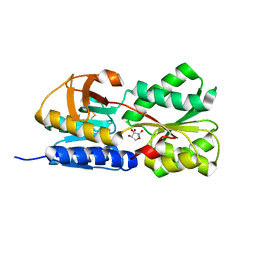 | | Crystal Structure of Salmonella typhimurium AI-2 receptor LsrB in complex with R-THMF | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
1TM2
 
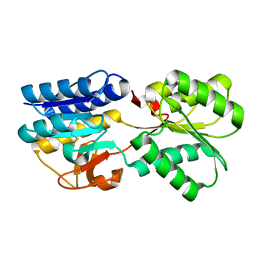 | | Crystal Structure of the apo form of the Salmonella typhimurium AI-2 receptor LsrB | | Descriptor: | sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-10 | | Release date: | 2004-09-28 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
3GND
 
 | |
3GLC
 
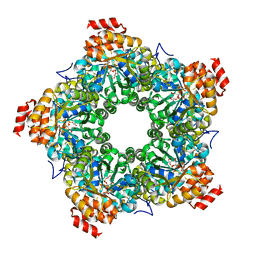 | |
3GKF
 
 | | Crystal Structure of E. coli LsrF | | Descriptor: | Aldolase lsrF | | Authors: | Miller, S.T, Diaz, Z.C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
Plos One, 4, 2009
|
|
6DSP
 
 | | LsrB from Clostridium saccharobutylicum in complex with AI-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein LsrB | | Authors: | Torcato, I.M, Xavier, K.B, Miller, S.T. | | Deposit date: | 2018-06-14 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of novel autoinducer-2 receptors inClostridiareveals plasticity in the binding site of the LsrB receptor family.
J.Biol.Chem., 294, 2019
|
|
4G0F
 
 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P6) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
4FXM
 
 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
1ZHH
 
 | | Crystal Structure of the Apo Form of Vibrio Harveyi LUXP Complexed with the Periplasmic Domain of LUXQ | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Autoinducer 2 sensor kinase/phosphatase luxQ, Autoinducer 2-binding periplasmic protein luxP | | Authors: | Neiditch, M.B, Federle, M.J, Miller, S.T, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Regulation of LuxPQ Receptor Activity by the Quorum-Sensing Signal Autoinducer-2.
Mol.Cell, 18, 2005
|
|
