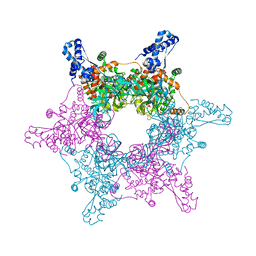
4R7Z
 
 | | PfMCM-AAA double-octamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 21, MAGNESIUM ION | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
4R7Y
 
 | | Crystal structure of an active MCM hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
8VON
 
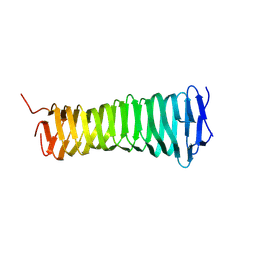 | |
8VOL
 
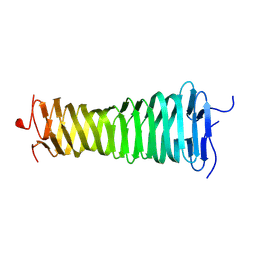 | |
8VOM
 
 | |
7TZ1
 
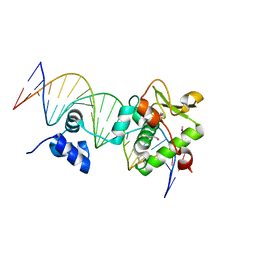 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
7R6R
 
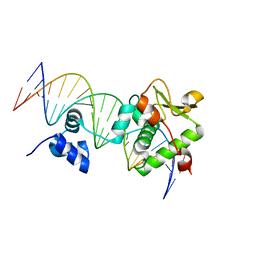 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
6VE6
 
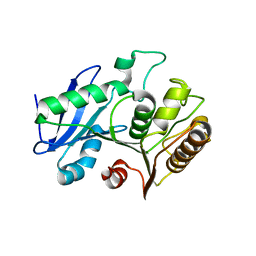 | | A structural characterization of poly(aspartic acid) hydrolase-1 from Sphingomonas sp. KT-1. | | Descriptor: | Poly(Aspartic acid) hydrolase-1 | | Authors: | Bolay, A.L, Salvo, H, Brambley, C.A, Yared, T.J, Miller, J.M, Wallen, J.R, Weiland, M.H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Structural Characterization of Sphingomonas sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
Acs Sustain Chem Eng, 2020
|
|
7LJI
 
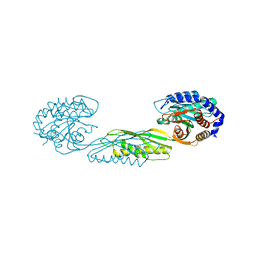 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7LJH
 
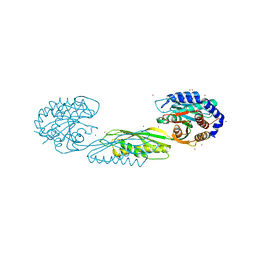 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Zn+2 bound | | Descriptor: | Poly(Aspartic acid) hydrolase, ZINC ION | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
