8IY8
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Feruloyl esterase | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
1OVT
 
 | |
4ZA2
 
 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
3IM0
 
 | | Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-08-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
4KEQ
 
 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4KEP
 
 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | Descriptor: | CITRATE ANION, GLYCEROL, VAL-1 | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
4TQV
 
 | | Crystal structure of a bacterial ABC transporter involved in the import of the acidic polysaccharide alginate | | Descriptor: | AlgM1, AlgM2, AlgS | | Authors: | Maruyama, Y, Itoh, T, Kaneko, A, Nishitani, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.504 Å) | | Cite: | Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Structure, 23, 2015
|
|
3DUV
 
 | | Crystal structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Haemophilus influenzae complexed with the substrate 3-deoxy-manno-octulosonate in the-configuration | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL | | Authors: | Yoon, H.J, Ku, M.J, Mikami, B, Suh, S.W. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Haemophilus influenzae complexed with the substrate 3-deoxy-manno-octulosonate in the beta-configuration.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1Q6C
 
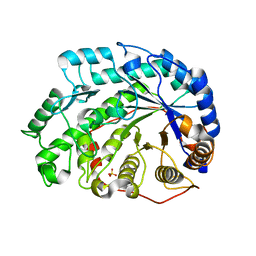 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1NFT
 
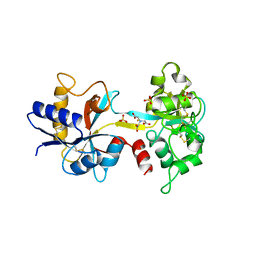 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1BFN
 
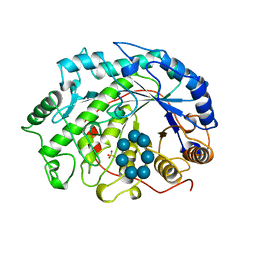 | | BETA-AMYLASE/BETA-CYCLODEXTRIN COMPLEX | | Descriptor: | BETA-AMYLASE, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), SULFATE ION | | Authors: | Adachi, M, Mikami, B, Katsube, T, Utsumi, S. | | Deposit date: | 1998-05-22 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of recombinant soybean beta-amylase complexed with beta-cyclodextrin.
J.Biol.Chem., 273, 1998
|
|
4U8G
 
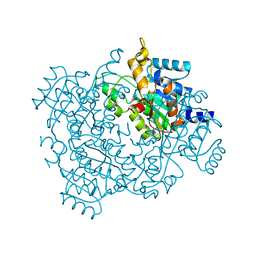 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1891 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4JY2
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4U8E
 
 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE
J.Biol.Chem., 290, 2015
|
|
4JY3
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
1NNT
 
 | |
3FKK
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
1Q1Y
 
 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
2GH4
 
 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
4TKZ
 
 | | Crystal structure of phosphotransferase system component EIIA from Streptococcus agalactiae | | Descriptor: | GLYCEROL, Putative uncharacterized protein gbs1890 | | Authors: | Nakamichi, Y, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-28 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphotransferase system component EIIA from Streptococcus agalactiae
To Be Published
|
|
4U8F
 
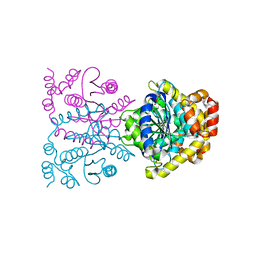 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase complexed with a tartrate | | Descriptor: | L(+)-TARTARIC ACID, Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
5GQP
 
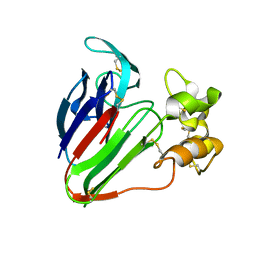 | | Thaumatin Structure at pH 8.0, orthorhombic type1 | | Descriptor: | Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Thaumatin Structure at pH 8.0, orthorhombic type1
To Be Published
|
|
3FGQ
 
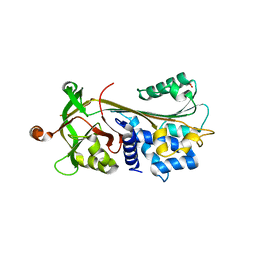 | | Crystal structure of native human neuroserpin | | Descriptor: | GLYCEROL, Neuroserpin | | Authors: | Takehara, S, Yang, X, Mikami, B, Onda, M. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability
J.Mol.Biol., 388, 2009
|
|
