6GCU
 
 | | MET receptor in complex with InlB internalin domain and DARPin A3A | | Descriptor: | DARPin A3A, Hepatocyte growth factor receptor, Internalin B | | Authors: | Meyer, T, Andres, F, Iamele, L, Gherardi, E, Pluckthun, A, Niemann, H.H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (6.001 Å) | | Cite: | Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking.
J.Mol.Biol., 431, 2019
|
|
6SMT
 
 | | S-enantioselective imine reductase from Mycobacterium smegmatis | | Descriptor: | (2S)-2-ethylhexan-1-ol, 1,2-ETHANEDIOL, 6-phosphogluconate dehydrogenase, ... | | Authors: | Meyer, T, Zumbraegel, N, Geerds, C, Groeger, H, Niemann, H.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of an S -enantioselective Imine Reductase from Mycobacterium Smegmatis .
Biomolecules, 10, 2020
|
|
2VCL
 
 | |
2VCK
 
 | |
2VGR
 
 | |
3IQD
 
 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
1Y57
 
 | | Structure of unphosphorylated c-Src in complex with an inhibitor | | Descriptor: | 4-[(4-METHYLPIPERAZIN-1-YL)METHYL]-N-{3-[(4-PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]PHENYL}BENZAMIDE, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Manley, P.W, Jahnke, W, Fabbro, D, Liebetanz, J, Meyer, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Crystal Structure of a c-Src Complex in an Active Conformation Suggests Possible Steps in c-Src Activation
Structure, 13, 2005
|
|
8PFH
 
 | | Crystal structure of the yeast septin complex Shs1-Cdc12-Cdc3-Cdc10 | | Descriptor: | CDC10 isoform 1, CDC12 isoform 1, Cell division control protein 3, ... | | Authors: | Grupp, B, Denkhaus, L, Gerhardt, S, Gronemeyer, T. | | Deposit date: | 2023-06-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | The structure of a tetrameric septin complex reveals a hydrophobic element essential for NC-interface integrity.
Commun Biol, 7, 2024
|
|
5AR1
 
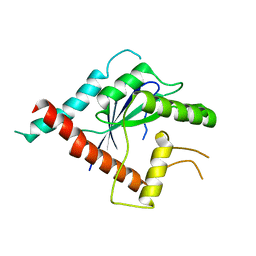 | | Crystal structure of Cdc11 from Saccharomyces cerevisiae | | Descriptor: | CELL DIVISION CONTROL PROTEIN 11 | | Authors: | Brausemann, A, Gerhardt, S, Schott, A.K, Einsle, O, Grosse-Berkenbusch, A, Johnsson, N, Gronemeyer, T. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Cdc11, a Septin Subunit from Saccharomyces Cerevisiae.
J.Struct.Biol., 193, 2016
|
|
4B0F
 
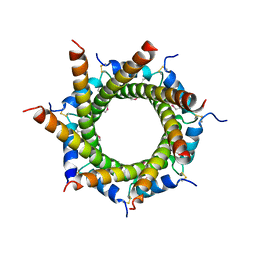 | | Heptameric core complex structure of C4b-binding (C4BP) protein from human | | Descriptor: | C4B-BINDING PROTEIN ALPHA CHAIN, CHLORIDE ION | | Authors: | Schmelz, S, Hofmeyer, T, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-07-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Arranged Sevenfold: Structural Insights Into the C-Terminal Oligomerization Domain of Human C4B-Binding Protein.
J.Mol.Biol., 425, 2013
|
|
