7KD3
 
 | |
6M7W
 
 | |
3R0J
 
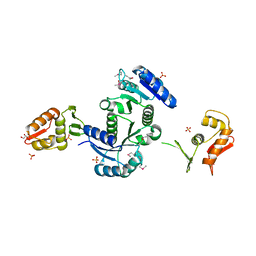 | | Structure of PhoP from Mycobacterium tuberculosis | | Descriptor: | POSSIBLE TWO COMPONENT SYSTEM RESPONSE TRANSCRIPTIONAL POSITIVE REGULATOR PHOP, R-1,2-PROPANEDIOL, SULFATE ION | | Authors: | Menon, S, Wang, S. | | Deposit date: | 2011-03-08 | | Release date: | 2011-07-13 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Response Regulator PhoP from Mycobacterium tuberculosis Reveals a Dimer through the Receiver Domain.
Biochemistry, 50, 2011
|
|
2W8M
 
 | |
5KBX
 
 | |
2VQC
 
 | | Structure of a DNA binding winged-helix protein, F-112, from Sulfolobus Spindle-shaped Virus 1. | | Descriptor: | HYPOTHETICAL 13.2 KDA PROTEIN | | Authors: | Menon, S.K, Kraft, P, Corn, G.J, Wiedenheft, B, Young, M.J, Lawrence, C.M. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Usage in Sulfolobus Spindle-Shaped Virus 1 and Extension to Hyperthermophilic Viruses in General.
Virology, 376, 2008
|
|
5DYM
 
 | | Crystal structure of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291 - CdPadR_0991 to 1.89 Angstrom resolution | | Descriptor: | PadR-family transcriptional regulator | | Authors: | Isom, C.E, Karr, E.A, Menon, S.K, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Crystal structure and DNA binding activity of a PadR family transcription regulator from hypervirulent Clostridium difficile R20291.
Bmc Microbiol., 16, 2016
|
|
4AAI
 
 | | THERMOSTABLE PROTEIN FROM HYPERTHERMOPHILIC VIRUS SSV-RH | | Descriptor: | ORF E73 | | Authors: | Schlenker, C, Goel, A, Tripet, B.P, Menon, S, Lawrence, C.M, Copie, V. | | Deposit date: | 2011-12-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of E73 from a Hyperthermophilic Archaeal Virus Identify the "Rh3" Domain, an Elaborated Ribbon-Helix- Helix Motif Involved in DNA Recognition.
Biochemistry, 51, 2012
|
|
2WBT
 
 | | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA | | Descriptor: | B-129, ZINC ION | | Authors: | Eilers, B.J, Menon, S, Windham, A.B, Kraft, P, Dlakic, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA
To be Published
|
|
1DB3
 
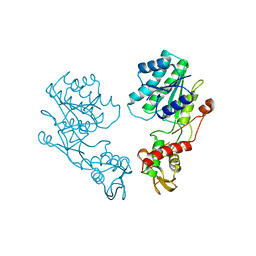 | | E.COLI GDP-MANNOSE 4,6-DEHYDRATASE | | Descriptor: | GDP-MANNOSE 4,6-DEHYDRATASE | | Authors: | Somoza, J.R, Menon, S, Somers, W.S, Sullivan, F.X. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose.
Structure Fold.Des., 8, 2000
|
|
1R42
 
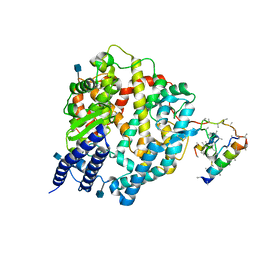 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R4L
 
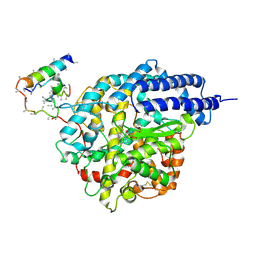 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
