1LRU
 
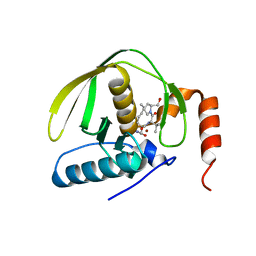 | | Crystal Structure of E.coli Peptide Deformylase Complexed with Antibiotic Actinonin | | 分子名称: | ACTINONIN, PEPTIDE DEFORMYLASE, SULFATE ION, ... | | 著者 | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | 登録日 | 2002-05-16 | | 公開日 | 2002-07-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1ZY0
 
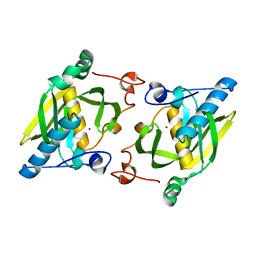 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-6000 | | 分子名称: | Peptide deformylase, mitochondrial, ZINC ION | | 著者 | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | 登録日 | 2005-06-09 | | 公開日 | 2005-09-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
7OWP
 
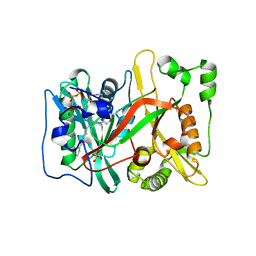 | | HsNMT1 in complex with both MyrCoA and ACE-G-(L-ORN)SFSKPR | | 分子名称: | ACE-GLY-ORN-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWR
 
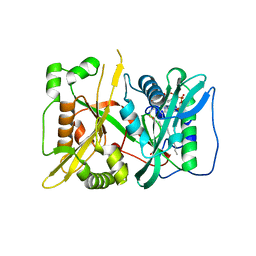 | | HsNMT1 in complex with both MyrCoA and peptide GGKSFSKPR | | 分子名称: | GLY-GLY-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWQ
 
 | |
7OWU
 
 | | HsNMT1 in complex with both CoA and Myr-ANCFSKPR peptide | | 分子名称: | ALA-ASN-CYS-PHE-SER-LYS-PRO-ARG, COENZYME A, GLYCEROL, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWO
 
 | | HsNMT1 in complex with both MyrCoA and N-acetylated KSFSKPR peptide | | 分子名称: | COENZYME A, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWN
 
 | | HsNMT1 in complex with both MyrCoA and peptide AKSFSKPR | | 分子名称: | ALA-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWM
 
 | | HsNMT1 in complex with both MyrCoA and HCPA substrate peptide GKQNSKLR | | 分子名称: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, Neuron-specific calcium-binding protein hippocalcin, ... | | 著者 | Dian, C, Giglione, C, Meinnel, T. | | 登録日 | 2021-06-18 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
3O3J
 
 | |
5MTC
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form I | | 分子名称: | NICKEL (II) ION, Putative uncharacterized protein orf60T, ZINC ION | | 著者 | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | 登録日 | 2017-01-09 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
3PN2
 
 | |
3PN3
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with inhibitor 21 | | 分子名称: | Peptide deformylase 1B, chloroplastic, ZINC ION, ... | | 著者 | Fieulaine, S, Meinnel, T, Giglione, C. | | 登録日 | 2010-11-18 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3PN4
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with actinonin (crystallized in PEG-550-MME) | | 分子名称: | ACTINONIN, Peptide deformylase 1B, chloroplastic, ... | | 著者 | Fieulaine, S, Meinnel, T, Giglione, C. | | 登録日 | 2010-11-18 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3PN6
 
 | |
5O9S
 
 | | HsNMT1 in complex with CoA and Myristoylated-GKSNSKLK octapeptide | | 分子名称: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | 著者 | Dian, C, Meinnel, T, Giglione, C. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5MTE
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T in complex with actinonin - crystal form II | | 分子名称: | ACTINONIN, NICKEL (II) ION, Putative uncharacterized protein orf60T, ... | | 著者 | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | 登録日 | 2017-01-09 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts.
Biochim. Biophys. Acta, 1866, 2018
|
|
5MTD
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form II | | 分子名称: | NICKEL (II) ION, Putative uncharacterized protein orf60T, TRIETHYLENE GLYCOL, ... | | 著者 | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | 登録日 | 2017-01-09 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
3PN5
 
 | |
5O9V
 
 | | HsNMT1 in complex with CoA and Myristoylated-GGCFSKPK octapeptide | | 分子名称: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | 著者 | Dian, C, Meinnel, T, Giglione, C. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5O9T
 
 | | HsNMT1 in complex with CoA and acetylated-NCFSKPK peptide | | 分子名称: | 1IP-CYS-PHE-SER-LYS-PRO-ARG, CHLORIDE ION, GLYCEROL, ... | | 著者 | Dian, C, Meinnel, T, Giglione, C. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5O9U
 
 | | HsNMT1 in complex with CoA and Myristoylated-GCSVSKKK octapeptide | | 分子名称: | COENZYME A, Calcineurin B-like protein 4, GLYCEROL, ... | | 著者 | Dian, C, Meinnel, T, Giglione, C. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5JF5
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT020 | | 分子名称: | (3R)-3-{3-[(2H-1,3-benzodioxol-5-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | 著者 | Fieulaine, S, Giglione, C, Meinnel, T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JEZ
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Ser | | 分子名称: | ACETATE ION, Met-Ala-Ser, Peptide deformylase, ... | | 著者 | Fieulaine, S, Giglione, C, Meinnel, T. | | 登録日 | 2016-04-19 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF7
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor SMP289 | | 分子名称: | 2-(3-benzyl-5-bromo-1H-indol-1-yl)-N-hydroxyacetamide, ACETATE ION, IMIDAZOLE, ... | | 著者 | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | 登録日 | 2016-04-19 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
