5ACC
 
 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
5AAV
 
 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
4A0G
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
1LUA
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 complexed with NADP | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
1N3Y
 
 | | Crystal structure of the alpha-X beta2 integrin I domain | | Descriptor: | Integrin alpha-X | | Authors: | Vorup-Jensen, T, Ostermeier, C, Shimaoka, M, Hommel, U, Springer, T.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and allosteric regulation of the alpha X beta 2 integrin I domain.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Z69
 
 | | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Ermler, U, Hagemeier, C.H, Thauer, R.K, Shima, S. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Protein Sci., 14, 2005
|
|
7BBR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCP
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with gluconate | | Descriptor: | D-gluconic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCO
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with D-foconate | | Descriptor: | (2R,3S,4S,5S)-2,3,4,5-tetrahydroxyhexanoic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with galactonate | | Descriptor: | L-galactonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCN
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with Xylonic acid | | Descriptor: | D-xylonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
4A0F
 
 | | Structure of selenomethionine substituted bifunctional DAPA aminotransferase-dethiobiotin synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0R
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to dethiobiotin (DTB). | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0H
 
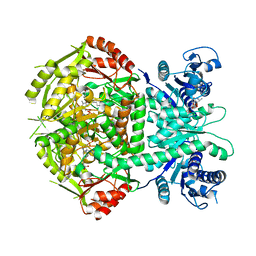 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to 7-keto 8-amino pelargonic acid (KAPA) | | Descriptor: | 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
2WD1
 
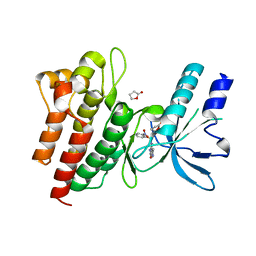 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1Y5Y
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 | | Descriptor: | CALCIUM ION, Formaldehyde-activating enzyme fae, SODIUM ION | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
1LU9
 
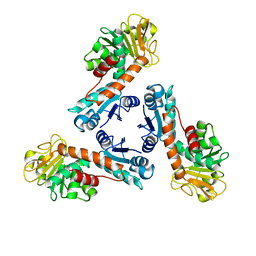 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1 | | Descriptor: | Methylene Tetrahydromethanopterin Dehydrogenase | | Authors: | Ermler, U, Hagemeier, C.H, Roth, A, Demmer, U, Grabarse, W, Warkentin, E, Vorholt, J.A. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1.
Structure, 10, 2002
|
|
5NFG
 
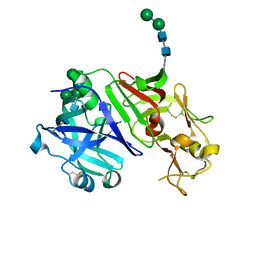 | | Structure of recombinant cardosin B from Cynara cardunculus | | Descriptor: | Procardosin-B,Procardosin-B, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pereira, P.J.B, Figueiredo, A.C, Manso, J.A, Almeida, C.M, Simoes, I. | | Deposit date: | 2017-03-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Functional and structural characterization of synthetic cardosin B-derived rennet.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
1Y60
 
 | | Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1 with bound 5,10-methylene tetrahydromethanopterin | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Formaldehyde-activating enzyme fae | | Authors: | Acharya, P, Goenrich, M, Hagemeier, C.H, Demmer, U, Vorholt, J.A, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme (Fae) from Methylobacterium extorquens AM1
J.Biol.Chem., 280, 2005
|
|
6FGB
 
 | | Human FcRn extra-cellular domain complexed with Fab fragment of Rozanolixizumab | | Descriptor: | 1519.g57- Heavy chain, 1519.g57- Light chain, Beta-2-microglobulin, ... | | Authors: | Sarkar, K, Ceska, T, Meier, C. | | Deposit date: | 2018-01-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Generation and characterization of a high affinity anti-human FcRn antibody, rozanolixizumab, and the effects of different molecular formats on the reduction of plasma IgG concentration.
MAbs, 10, 2018
|
|
1U6J
 
 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.4A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2V9C
 
 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
2B0J
 
 | | The crystal structure of the apoenzyme of the iron-sulfur-cluster-free hydrogenase (Hmd) | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase | | Authors: | Pilak, O, Mamat, B, Vogt, S, Hagemeier, C.H, Thauer, R.K, Shima, S, Vonrhein, C, Warkentin, E, Ermler, U. | | Deposit date: | 2005-09-14 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Apoenzyme of the Iron-Sulphur Cluster-free Hydrogenase
J.Mol.Biol., 358, 2006
|
|
1U6I
 
 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U6K
 
 | | TLS refinement of the structure of Se-methionine labelled Coenzyme f420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) from Methanopyrus kandleri | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
