3DBK
 
 | | Pseudomonas aeruginosa elastase with phosphoramidon | | Descriptor: | CALCIUM ION, Elastase, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | McKay, D.B, Overgaard, M.T. | | Deposit date: | 2008-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Elastase of Pseudomonas aeruginosa Complexed with Phosphoramidon
To be Published
|
|
4NBO
 
 | | Human steroid receptor RNA activator protein carboxy-terminal domain | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Mckay, D.B, Xi, L, Barthel, K.K.B, Cech, T.C. | | Deposit date: | 2013-10-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structure and function of steroid receptor RNA activator protein, the proposed partner of SRA noncoding RNA.
J.Mol.Biol., 426, 2014
|
|
3INK
 
 | |
2G0C
 
 | |
2HJV
 
 | |
1IKQ
 
 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKP
 
 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1U4G
 
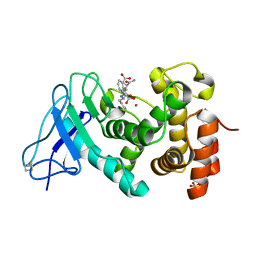 | | Elastase of Pseudomonas aeruginosa with an inhibitor | | Descriptor: | CALCIUM ION, Elastase, N-(1-CARBOXY-3-PHENYLPROPYL)PHENYLALANYL-ALPHA-ASPARAGINE, ... | | Authors: | Bitto, E, McKay, D.B. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elastase of Pseudomonas aeruginosa with an inhibitor
To be Published, 2004
|
|
2PV3
 
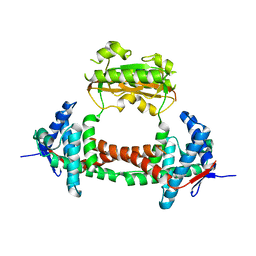 | |
3MOJ
 
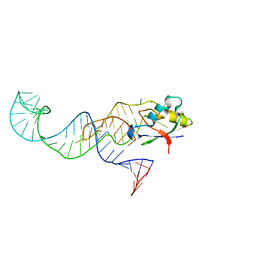 | |
2PV2
 
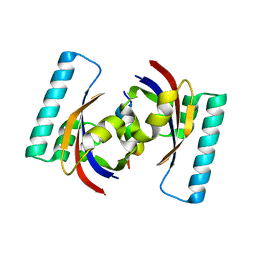 | |
2PV1
 
 | |
429D
 
 | |
3HSC
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ATPASE FRAGMENT OF A 70K HEAT-SHOCK COGNATE PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 7OKD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Deluca-Flaherty, C.R, Mckay, D.B. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein.
Nature, 346, 1990
|
|
2BUP
 
 | | T13G Mutant of the ATPASE fragment of Bovine HSC70 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sousa, M.C, Mckay, D.B. | | Deposit date: | 1998-09-08 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
1KAX
 
 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71M MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1KAZ
 
 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71E MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1G41
 
 | | CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1KYI
 
 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | Authors: | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2002-02-04 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
1OFI
 
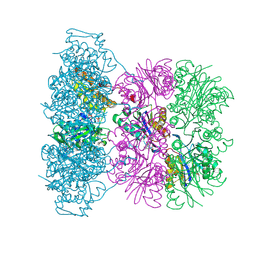 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFH
 
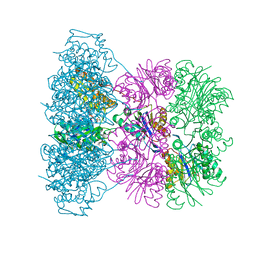 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1NGD
 
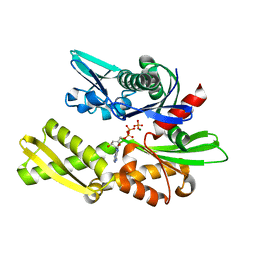 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGJ
 
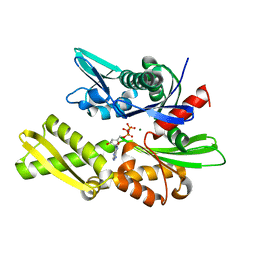 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGC
 
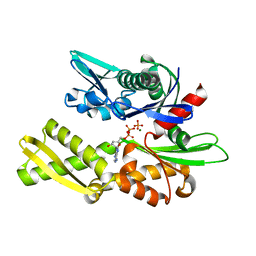 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NGI
 
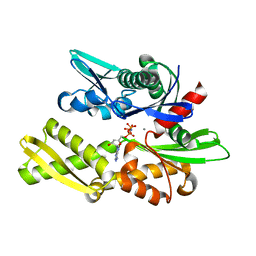 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | CALCIUM ION, HEAT-SHOCK COGNATE 70 kD PROTEIN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
