242D
 
 | | MAD PHASING STRATEGIES EXPLORED WITH A BROMINATED OLIGONUCLEOTIDE CRYSTAL AT 1.65 A RESOLUTION. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(CBR)P*G)-3') | | Authors: | Peterson, M.R, Harrop, S.J, McSweeney, S.M, Leonard, G.A, Thompson, A.W, Hunter, W.N, Helliwell, J.R. | | Deposit date: | 1996-06-20 | | Release date: | 1996-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MAD Phasing Strategies Explored with a Brominated Oligonucleotide Crystal at 1.65A Resolution.
J.Synchrotron Radiat., 3, 1996
|
|
1HJ9
 
 | | Atomic resolution structures of trypsin provide insight into structural radiation damage | | Descriptor: | ANILINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, McSweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-10 | | Release date: | 2002-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6C5Y
 
 | | Crystal structure of thaumatin from microcrystals | | Descriptor: | Thaumatin-1 | | Authors: | Guo, G, Fuchs, M, Shi, W, Skinner, J, Berman, E, Ogata, C.M, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sample manipulation and data assembly for robust microcrystal synchrotron crystallography.
IUCrJ, 5, 2018
|
|
6PBR
 
 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | Authors: | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4JCV
 
 | |
4UNF
 
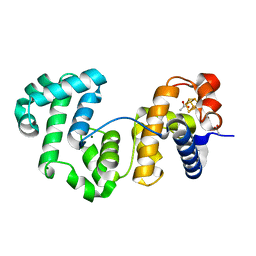 | | Crystal structure of Deinococcus radiodurans Endonuclease III-1 | | Descriptor: | ACETATE ION, ENDONUCLEASE III-1, IRON/SULFUR CLUSTER, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
8F6T
 
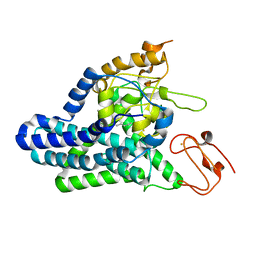 | | Cryo-EM structure of alkane 1-monooxygenase AlkB-AlkG complex from Fontimonas thermophila | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION | | Authors: | Chai, J, Guo, G, McSweeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for enzymatic terminal C-H bond functionalization of alkanes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4UOB
 
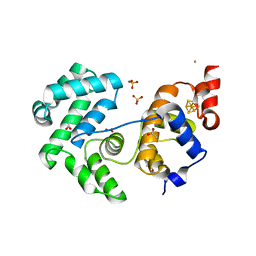 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
7MHF
 
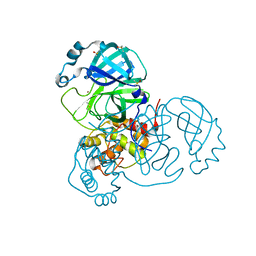 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHL
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
6TGT
 
 | |
6TB5
 
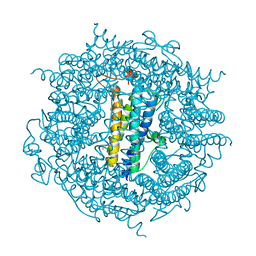 | | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h). | | Descriptor: | CALCIUM ION, DNA protection during starvation protein 2, FE (III) ION | | Authors: | Cuypers, M.G, McSweeney, S, Romao, C.V, Mitchell, E.P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h).
To Be Published
|
|
7SEU
 
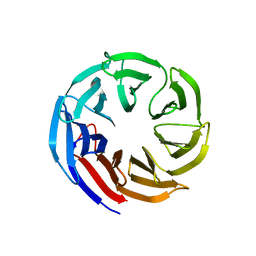 | | Crystal structure of E coli contaminant protein YncE co-purified with a plant protein | | Descriptor: | PQQ-dependent catabolism-associated beta-propeller protein | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
8ENA
 
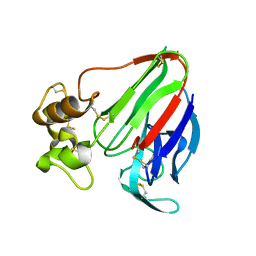 | | Thaumatin native-SAD structure determined at 5 keV with a helium environmet | | Descriptor: | Thaumatin-1 | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
8EN9
 
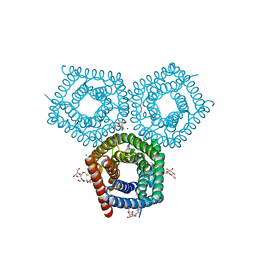 | | TehA native-SAD structure determined at 5 keV with a helium environment | | Descriptor: | CHLORIDE ION, SODIUM ION, Tellurite resistance protein TehA homolog, ... | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
6O8A
 
 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
7SEV
 
 | | Crystal structure of E coli contaminant protein YadF co-purified with a plant protein | | Descriptor: | Carbonic anhydrase 2, POTASSIUM ION, ZINC ION | | Authors: | Chai, L, Zhu, P, Chai, J, Pang, C, Andi, B, McsWeeney, S, Shanklin, J, Liu, Q. | | Deposit date: | 2021-10-01 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
Crystals, 11, 2021
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | Deposit date: | 2022-01-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
6W8R
 
 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8T
 
 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8S
 
 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
2JG1
 
 | | STRUCTURE OF Staphylococcus aureus D-TAGATOSE-6-PHOSPHATE KINASE with cofactor and substrate | | Descriptor: | 6-O-phosphono-beta-D-tagatofuranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Miallau, L, Hunter, W.N, McSweeney, S.M, Leonard, G.A. | | Deposit date: | 2007-02-07 | | Release date: | 2007-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase Implicate Domain Motions in Specificity and Mechanism.
J.Biol.Chem., 282, 2007
|
|
2JGV
 
 | | STRUCTURE OF Staphylococcus aureus D-TAGATOSE-6-PHOSPHATE KINASE in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TAGATOSE-6-PHOSPHATE KINASE | | Authors: | Miallau, L, Hunter, W.N, McSweeney, S.M, Leonard, G.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Staphylococcus Aureus D-Tagatose-6-Phosphate Kinase Implicate Domain Motions in Specificity and Mechanism.
J.Biol.Chem., 282, 2007
|
|
