1QVA
 
 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
1EZM
 
 | |
1N5B
 
 | |
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1M5Y
 
 | |
1KAP
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF | | Descriptor: | ALKALINE PROTEASE, CALCIUM ION, TETRAPEPTIDE (GLY SER ASN SER), ... | | Authors: | Baumann, U, Wu, S, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1995-06-08 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif.
EMBO J., 12, 1993
|
|
1G3K
 
 | |
1FUU
 
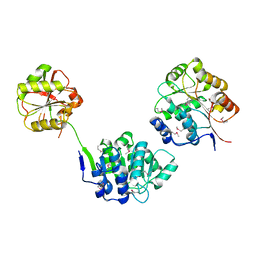 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FUK
 
 | |
