1QOV
 
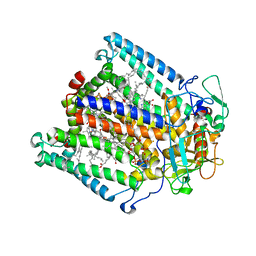 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M260 REPLACED WITH TRP (CHAIN M, A260W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | McAuley, K.E, Fyfe, P.K, Ridge, J.P, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1999-11-17 | | Release date: | 1999-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Details of an Interaction between Cardiolipin and an Integral Membrane Protein
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
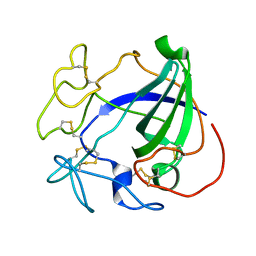
1HD5
 
 | |
5FFN
 
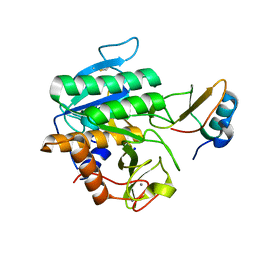 | | Complex of subtilase SubTY from Bacillus sp. TY145 with chymotrypsin inhibitor CI2A | | Descriptor: | CALCIUM ION, Enzyme subtilase SubTY from Bacillus sp. TY145, SODIUM ION, ... | | Authors: | McAuley, K.E, Svendsen, A, Oestergaard, P.R, Dohnalek, J, Wilson, K.S. | | Deposit date: | 2015-12-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
1EB6
 
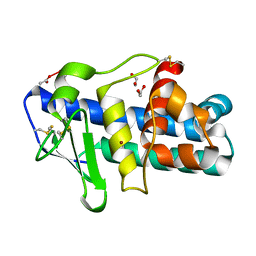 | | Deuterolysin from Aspergillus oryzae | | Descriptor: | 1,2-ETHANEDIOL, NEUTRAL PROTEASE II, ZINC ION | | Authors: | McAuley, K.E, Jia-Xing, Y, Dodson, E.J, Lehmbeck, J, Ostergaard, P.R, Wilson, K.S. | | Deposit date: | 2001-07-19 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Quick Solution: Ab Initio Structure Determination of a 19 kDa Metalloproteinase Using Acorn
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1UWC
 
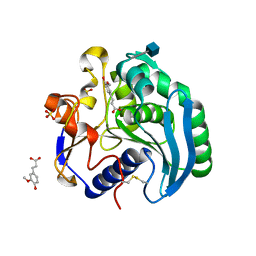 | | Feruloyl esterase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, FERULOYL ESTERASE A, ... | | Authors: | McAuley, K.E, Svendsen, A, Patkar, S.A, Wilson, K.S. | | Deposit date: | 2004-02-03 | | Release date: | 2004-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of a Feruloyl Esterase from Aspergillus Niger
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UZA
 
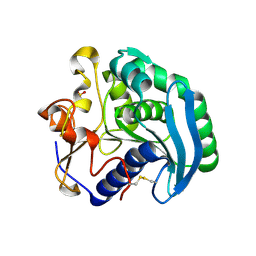 | | Crystallographic structure of a feruloyl esterase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FERULOYL ESTERASE A, SULFATE ION | | Authors: | McAuley, K.E, Svendsen, A, Patkar, S.A, Wilson, K.S. | | Deposit date: | 2004-03-05 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Feruloyl Esterase from Aspergillus Niger
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7PB3
 
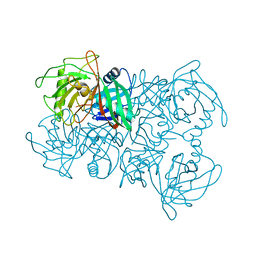 | | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
1E6D
 
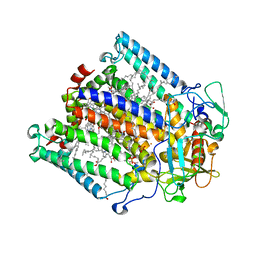 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH TRP M115 REPLACED WITH PHE (CHAIN M, WM115F) PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ridge, J.P, Fyfe, P.K, McAuley, K.E, Van Brederode, M.E, Robert, B, Van Grondelle, R, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2000-08-11 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Examination of How Structural Changes Can Affect the Rate of Electron Transfer in a Mutated Bacterial Photoreaction Centre
Biochem.J., 351, 2000
|
|
8A4R
 
 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic orthorhombic data at 3.59 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
8A3F
 
 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic tetragonal data at 3.15 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
1E14
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) AND GLY M203 REPLACED WITH ASP (CHAIN M, GM203D) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Ridge, J.P, McAuley, K.E, Cogdell, R.J, Isaacs, N.W, Jones, M.R. | | Deposit date: | 2000-04-18 | | Release date: | 2000-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Consequences of the Replacement of Glycine M203 with Aspartic Acid in the Reaction Center from Rhodobacter Sphaeroides.
Biochemistry, 39, 2000
|
|
3KSA
 
 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (cleaved form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', 5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
3LTN
 
 | | Inhibitor-stabilized topoisomerase IV-DNA cleavage complex (S. pneumoniae) | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2010-02-16 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
3KSB
 
 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (re-sealed form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*TP*GP*AP*CP*TP*AP*TP*GP*CP*AP*CP*GP*TP*AP*AP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*TP*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', DNA topoisomerase 4 subunit A, ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
5I3D
 
 | | Sulfolobus solfataricus beta-glycosidase - E387Y mutant | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, Beta-galactosidase | | Authors: | Iglesias-Fernandez, J, Hancock, S.M, Lee, S.S, McAuley, K.E, Fordham-Skelton, A, Rovira, C, Davis, B.D. | | Deposit date: | 2016-02-10 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A front-face 'SNi synthase' engineered from a retaining 'double-SN2' hydrolase.
Nat. Chem. Biol., 13, 2017
|
|
3FOE
 
 | | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-8-chloro-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*AP*CP*CP*AP*AP*GP*GP*TP*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Laponogov, I, Sohi, M.K, Veselkov, D.A, Pan, X.-S, Sawhney, R, Thompson, A.W, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2008-12-30 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Nat.Struct.Mol.Biol., 16, 2009
|
|
3FOF
 
 | | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*AP*CP*CP*AP*AP*GP*GP*TP*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Laponogov, I, Sohi, M.K, Veselkov, D.A, Pan, X.-S, Sawhney, R, Thompson, A.W, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2008-12-30 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Nat.Struct.Mol.Biol., 16, 2009
|
|
3RAF
 
 | | Quinazolinedione-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibitor-stabilised cleavage complexes of topoisomerase IIa: structural analysis of drug-dependent inter- and intramolecular interactions
To be Published
|
|
3RAE
 
 | | Quinolone(Levofloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3RAD
 
 | | Quinolone(Clinafloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', 5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*G)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
