3U93
 
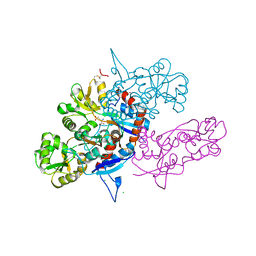 | |
8U7I
 
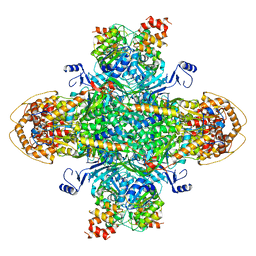 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
4WXJ
 
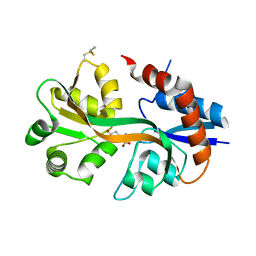 | | Drosophila muscle GluRIIB complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor IIB,Glutamate receptor IIB | | Authors: | Dharkar, P, Mayer, M.L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UQJ
 
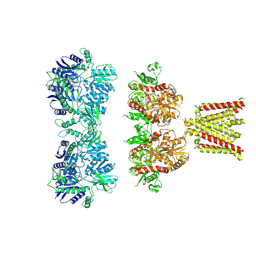 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQ6
 
 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQQ
 
 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
5ICT
 
 | |
4YKP
 
 | |
4YKI
 
 | |
4YKJ
 
 | |
4YKK
 
 | |
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
5DT6
 
 | |
5DTB
 
 | |
5KUF
 
 | | GluK2EM with 2S,4R-4-methylglutamate | | Descriptor: | 2S,4R-4-METHYLGLUTAMATE, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5KUH
 
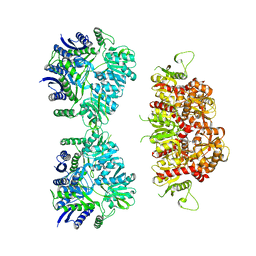 | | GluK2EM with LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5EHM
 
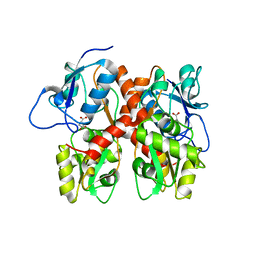 | |
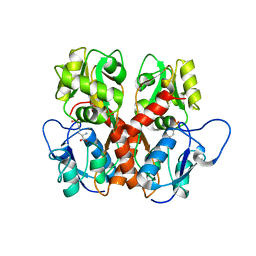
5EHS
 
 | |
1MQJ
 
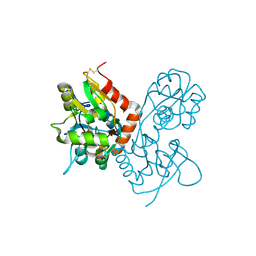 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MM7
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Quisqualate in a Zinc Crystal Form at 1.65 Angstroms Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Activation and Selectivity in a Ligand-Gated Ion Channel: Structural and Functional Studies of GluR2 and Quisqualate
Biochemistry, 41, 2003
|
|
1MM6
 
 | | crystal structure of the GluR2 ligand binding core (S1S2J) in complex with quisqualate in a non zinc crystal form at 2.15 angstroms resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, GLYCEROL, ... | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of activation and selectivity in a ligand-gated ion channel: Structural and functional studies of GluR2 and quisqualate
Biochemistry, 41, 2002
|
|
1MQG
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
