2MPV
 
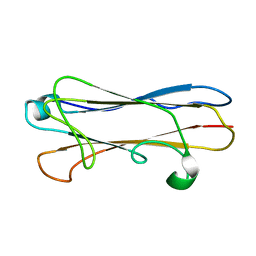 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
8AG0
 
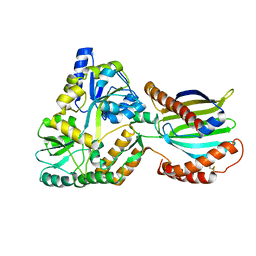 | | Crystal structure of mutant PRELID3a-TRIAP1 complex - R53E | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3A, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Milara, X, Perez-Dorado, J.I, Matthews, S.J. | | Deposit date: | 2022-07-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An intermolecular hydrogen bonded network in the PRELID-TRIAP protein family plays a role in lipid sensing.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
4XZS
 
 | | Crystal Structure of TRIAP1-MBP fusion | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Abid-Ali, F, Perez-Dorado, I, Matthews, S.J. | | Deposit date: | 2015-02-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
6TXT
 
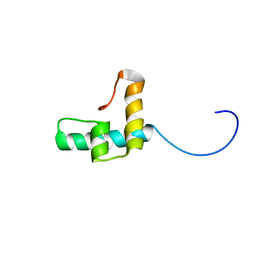 | |
5LG9
 
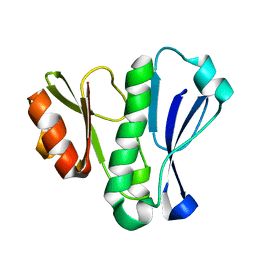 | |
1HD9
 
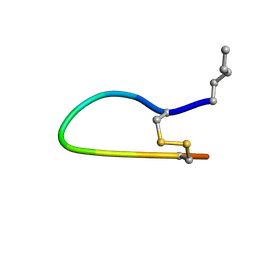 | | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif | | Descriptor: | BOWMAN-BIRK INHIBITOR DERIVED PEPTIDE | | Authors: | Brauer, A.B.E, Kelly, G, McBride, J.D, Cooke, R.M, Matthews, S.J, Leatherbarrow, R.J. | | Deposit date: | 2000-11-13 | | Release date: | 2001-03-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif
J.Mol.Biol., 306, 2001
|
|
6Y1H
 
 | |
6I3Y
 
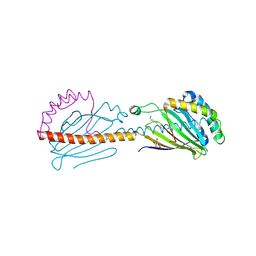 | | Crystal structure of the human mitochondrial PRELID1K58V-TRIAP1 complex with PS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, PRELI domain-containing protein 1, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I4Y
 
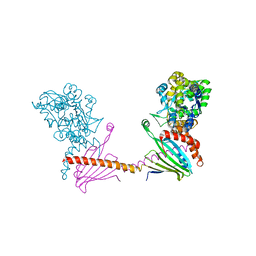 | | X-ray structure of the human mitochondrial PRELID3b-TRIAP1 complex | | Descriptor: | Maltose transport system, substrate-binding protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3B, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I3V
 
 | | x-ray structure of the human mitochondrial PRELID1 in complex with TRIAP1 | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, PRELI domain-containing protein 1, ... | | Authors: | Berry, J.L, Miliara, X, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
8C4A
 
 | |
5O65
 
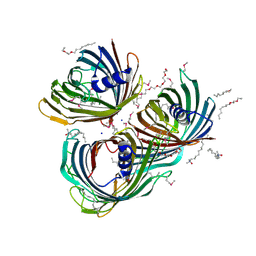 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O67
 
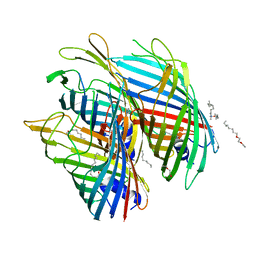 | | Crystal structure of the FapF polypeptide transporter - F103A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O68
 
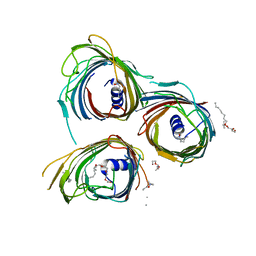 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF - R157A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
6FI7
 
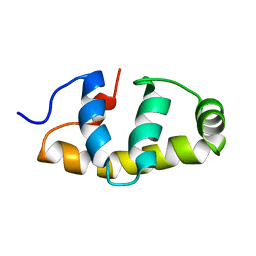 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FUE
 
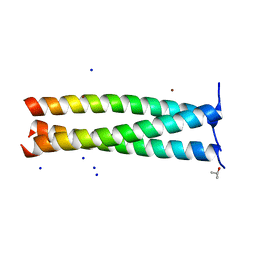 | | Periplasmic coiled coil domain of the FapF amyloid transporter | | Descriptor: | FapF, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Rouse, S.L, Stylianou, F, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The FapF Amyloid Secretion Transporter Possesses an Atypical Asymmetric Coiled Coil.
J. Mol. Biol., 430, 2018
|
|
3QS2
 
 | |
3QS3
 
 | |
3UIY
 
 | |
3RGU
 
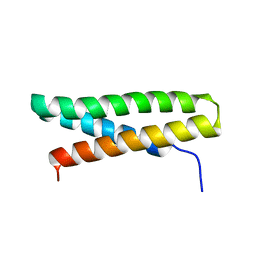 | | Structure of Fap-NRa at pH 5.0 | | Descriptor: | Fimbriae-associated protein Fap1, alpha-D-glucopyranose | | Authors: | Garnett, J.A, Matthews, S.J. | | Deposit date: | 2011-04-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation.
Biochem.Biophys.Res.Commun., 417, 2012
|
|
3UIZ
 
 | |
1GM2
 
 | |
4CL1
 
 | | The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors | | Descriptor: | NON-STRUCTURAL PROTEIN 5A, SULFATE ION, ZINC ION | | Authors: | Lambert, S.M, Langley, D.R, Garnett, J.A, Angell, R, Hedgethorne, K, Meanwell, N.A, Matthews, S.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of Ns5A Domain 1 from Genotype 1A Reveals New Clues to the Mechanism of Action for Dimeric Hcv Inhibitors.
Protein Sci., 23, 2014
|
|
2BEY
 
 | |
5CIV
 
 | | Sibling Lethal Factor Precursor - DfsB | | Descriptor: | Sibling bacteriocin | | Authors: | Taylor, J.D, Matthews, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.384 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
