9AX8
 
 | | 70S initiation complex (tRNA-fMet M1, initiation factor 2 + CUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
9AX7
 
 | | 70S initiation complex (tRNA-fMet M1 + CUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
9CG6
 
 | | 70S initiation complex (tRNA-fMet M1 + GUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-06-28 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
9CG5
 
 | | 70S initiation complex (tRNA-fMet M1 + UUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-06-28 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
9CG7
 
 | | 70S initiation complex (tRNA-fMet M1 + AUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-06-28 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
1TBZ
 
 | | HUMAN THROMBIN WITH ACTIVE SITE N-METHYL-D PHENYLALANYL-N-[5-(AMINOIMINOMETHYL)AMINO]-1-{{BENZOTHIAZOLYL)CARBONYL] BUTYL]-L-PROLINAMIDE TRIFLUROACETATE AND EXOSITE-HIRUGEN | | Descriptor: | ALPHA-THROMBIN, D-phenylalanyl-N-{(1S)-1-[(S)-1,3-benzothiazol-2-yl(hydroxy)methyl]-4-carbamimidamidobutyl}-L-prolinamide, HIRUGEN, ... | | Authors: | Matthews, J.H, Krishnan, R, Costanzo, M.J, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thrombin with thiazole-containing inhibitors: probes of the S1' binding site.
Biophys.J., 71, 1996
|
|
5I2A
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diol-dehydratase | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
5I2G
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | Diol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
5I3T
 
 | | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans | | Descriptor: | 1,3-BUTANEDIOL, CHLORIDE ION, Linalool dehydratase/isomerase, ... | | Authors: | LaMattina, J.W, Carlock, M, Koch, D.J, Lanzilotta, W.N. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans
To Be Published
|
|
5FFQ
 
 | | ChuY: An Anaerobillin Reductase from Escherichia coli O157:H7 | | Descriptor: | 1,4-BUTANEDIOL, PHOSPHATE ION, ShuY-like protein | | Authors: | LaMattina, J.W, Reedy, A.N, Uy, K.G, Lanzilotta, W.N. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Radical new paradigm for heme degradation in Escherichia coli O157:H7.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4MTJ
 
 | | Structure of the b12-independent glycerol dehydratase with 1,2-propanediol bound | | Descriptor: | B12-independent glycerol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J, Wright, A.V, Demick, J, Soucaille, P, Lanzilotta, W.N. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | When Computational Chemistry and Modern Software Get It Right; New Insight Into the Mechanism of a Glycyl Radical Enzyme
To be Published
|
|
1A4W
 
 | | CRYSTAL STRUCTURES OF THROMBIN WITH THIAZOLE-CONTAINING INHIBITORS: PROBES OF THE S1' BINDING SITE | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Matthews, J.H, Krishnan, R, Costanzo, M.J, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1998-02-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of thrombin with thiazole-containing inhibitors: probes of the S1' binding site.
Biophys.J., 71, 1996
|
|
4QGS
 
 | |
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
2DQ5
 
 | | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | | Descriptor: | Midline-1, ZINC ION | | Authors: | Massiah, M.A, Matts, J.A.B, Short, K.M, Simmons, B.N, Singireddy, S, Zou, J, Cox, T.C. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold
J.Mol.Biol., 369, 2007
|
|
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
6PTL
 
 | |
1J2O
 
 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1RUT
 
 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
2DFY
 
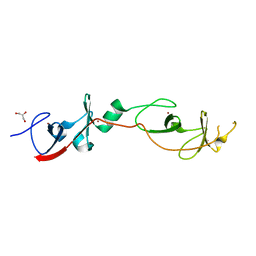 | | Crystal structure of a cyclized protein fusion of LMO4 LIM domains 1 and 2 with the LIM interacting domain of LDB1 | | Descriptor: | GLYCEROL, ZINC ION, fusion protein of LIM domain transcription factor LMO4 and LIM domain-binding protein 1 | | Authors: | Jeffries, C.M.J, Graham, S.C, Collyer, C.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of a binary protein complex by intein-mediated cyclization
Protein Sci., 15, 2006
|
|
9BPO
 
 | |
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1U85
 
 | |
1SRK
 
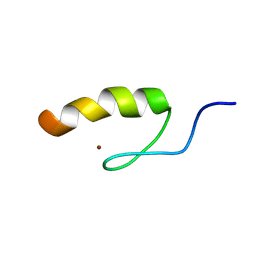 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
