7P1K
 
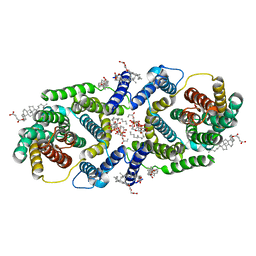 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
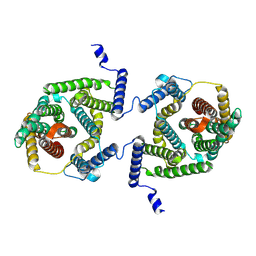 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1J
 
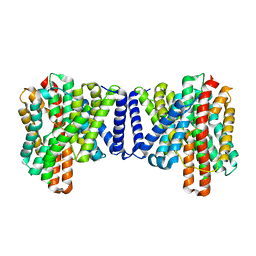 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5AZ6
 
 | |
5AZ7
 
 | |
5AZ8
 
 | |
5AZ9
 
 | |
5AZA
 
 | |
3WOV
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PaAglB-L, Q9V250_PYRAB, PAB2202) from Pyrococcus abyssi | | Descriptor: | CALCIUM ION, Oligosaccharyl transferase | | Authors: | Matsuoka, R, Nyirenda, J, Maita, N, Kohda, D. | | Deposit date: | 2014-01-05 | | Release date: | 2014-01-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PaAglB-L, Q9V250_PYRAB, PAB2202) from Pyrococcus abyssi
To be Published
|
|
6Z3Z
 
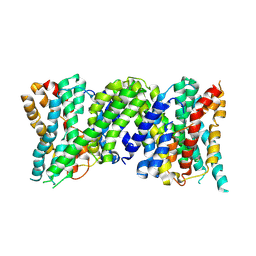 | |
7S24
 
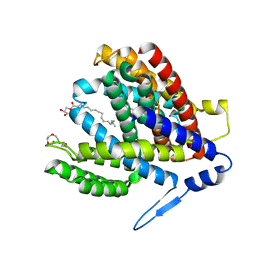 | | Crystal structure of the Na+/H+ antiporter NhaA at pH 6.5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Na(+)/H(+) antiporter NhaA, PENTAETHYLENE GLYCOL | | Authors: | Drew, D, Brock, J, Uzdavinys, P, Matsuoka, R. | | Deposit date: | 2021-09-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Na + /H + antiporter NhaA at active pH reveals the mechanistic basis for pH sensing.
Nat Commun, 13, 2022
|
|
6Z3Y
 
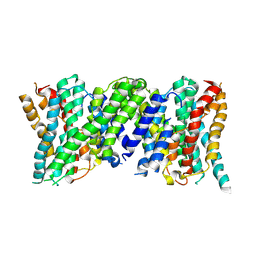 | |
5UQY
 
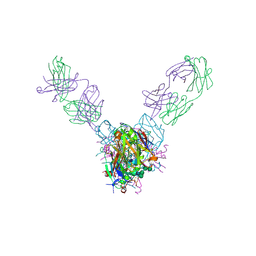 | | Crystal structure of Marburg virus GP in complex with the human survivor antibody MR78 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP1, ... | | Authors: | Hashiguchi, T, Fusco, M.L, Hastie, K.M, Bomholdt, Z.A, Lee, J.E, Flyak, A.I, Matsuoka, R, Kohda, D, Yanagi, Y, Hammel, M, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160, 2015
|
|
6RW3
 
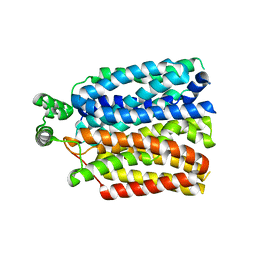 | | The molecular basis for sugar import in malaria parasites. | | Descriptor: | Hexose transporter 1, alpha-D-glucopyranose, beta-D-glucopyranose | | Authors: | Qureshi, A, Matsuoka, R, Brock, J, Drew, D. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The molecular basis for sugar import in malaria parasites.
Nature, 578, 2020
|
|
8QSN
 
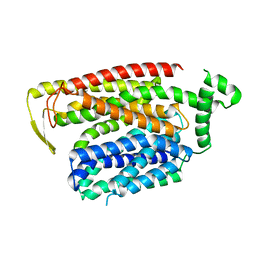 | |
8QSM
 
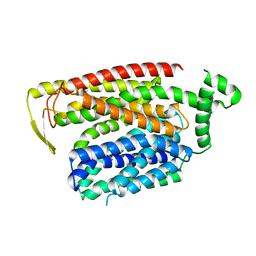 | |
8QSL
 
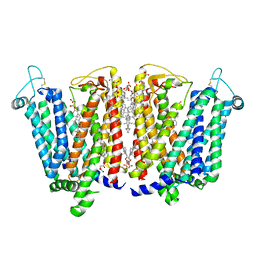 | | Cryo-EM structure of human SLC15A4 dimer in outward open state in LMNG | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Rehan, S, Matsuoka, R, Jazayeri, A, Duerr, K.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of human SLC15A4 dimer in outward open state in LMNG
To Be Published
|
|
8QSK
 
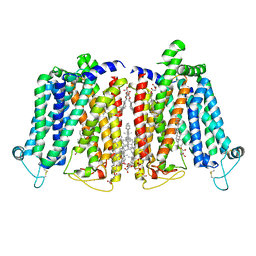 | | Cryo-EM structure of human SLC15A4 dimer in outward open state in MSP1D1 nanodisc | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Rehan, S, Matsuoka, R, Jazayeri, A, Duerr, K.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of human SLC15A4 dimer in outward open state in MSP1D1 nanodisc
To Be Published
|
|
5B2D
 
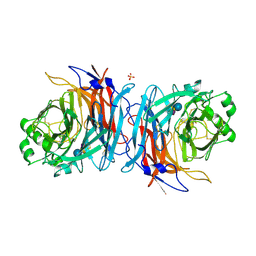 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8PXB
 
 | |
5ZFU
 
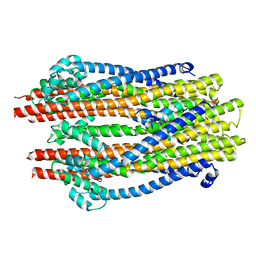 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5YXW
 
 | | Crystal structure of the prefusion form of measles virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZD
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor peptide (FIP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZFP
 
 | |
5YZC
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
