1WCW
 
 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
3WCK
 
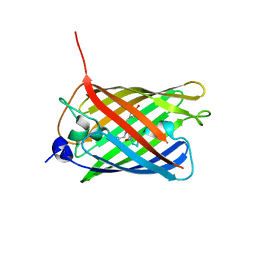 | | Crystal structure of monomeric photosensitizing fluorescent protein, Supernova | | Descriptor: | Monomeric photosenitizing fluorescent protein supernova | | Authors: | Sakai, N, Matsuda, T, Takemoto, K, Nagai, T. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation
Sci Rep, 3, 2013
|
|
3WIN
 
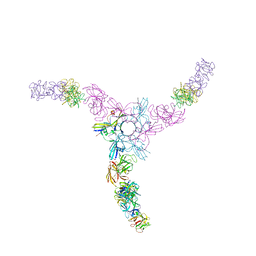 | | Clostridium botulinum Hemagglutinin | | Descriptor: | 17 kD hemagglutinin component, HA1, HA3 | | Authors: | Amatsu, S, Sugawara, Y, Matsumura, T, Fujinaga, Y, Kitadokoro, K. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Clostridium botulinum Whole Hemagglutinin Reveals a Huge Triskelion-shaped Molecular Complex
J.Biol.Chem., 288, 2013
|
|
1RTU
 
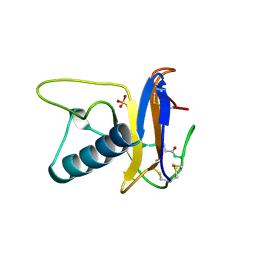 | | USTILAGO SPHAEROGENA RIBONUCLEASE U2 | | Descriptor: | RIBONUCLEASE U2, SULFATE ION | | Authors: | Noguchi, S, Satow, Y, Uchida, T, Sasaki, C, Matsuzaki, T. | | Deposit date: | 1995-05-12 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ustilago sphaerogena ribonuclease U2 at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
7CP7
 
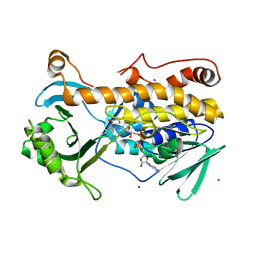 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
5XGZ
 
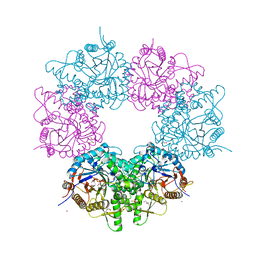 | | Metagenomic glucose-tolerant glycosidase | | Descriptor: | Beta-glycosidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Watanabe, M, Matsuzawa, T, Yaoi, K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Improved thermostability of a metagenomic glucose-tolerant beta-glycosidase based on its X-ray crystal structure.
Appl.Microbiol.Biotechnol., 101, 2017
|
|
1UC9
 
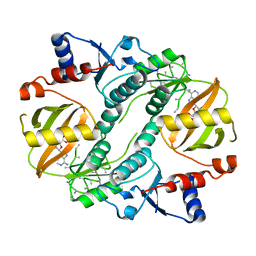 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
6KYE
 
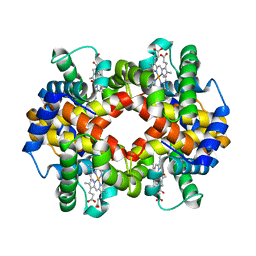 | | The crystal structure of recombinant human adult hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Funaki, R, Okamoto, W, Endo, C, Morita, Y, Komatsu, T. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Genetically engineered haemoglobin wrapped covalently with human serum albumins as an artificial O2carrier.
J Mater Chem B, 8, 2020
|
|
3WZU
 
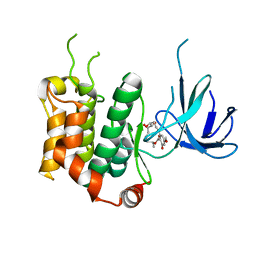 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
3AUJ
 
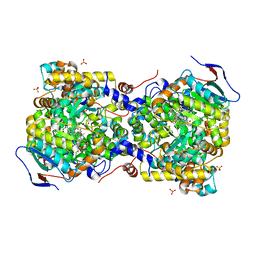 | | Structure of diol dehydratase complexed with glycerol | | Descriptor: | CALCIUM ION, COBALAMIN, Diol dehydrase alpha subunit, ... | | Authors: | Yamanishi, M, Kinoshita, K, Fukuoka, M, Shibata, T, Tobimatsu, T, Toraya, T. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols
Febs J., 279, 2012
|
|
2ZWB
 
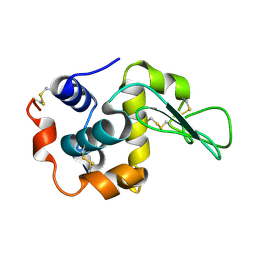 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
1S2H
 
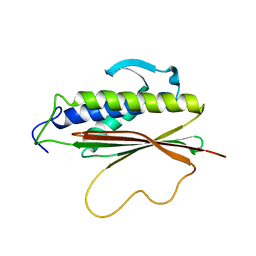 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2ZDY
 
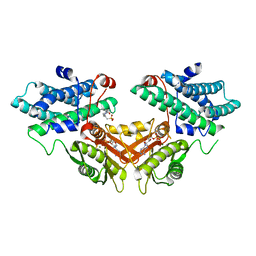 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3W5W
 
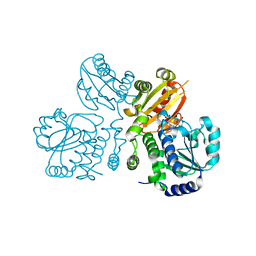 | | Mn2+-GMP complex of nanoRNase (Nrn) from Bacteroides fragilis | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Putative exopolyphosphatase-related protein | | Authors: | Uemura, Y, Nakagawa, N, Wakamatsu, T, Montelione, G.T, Hunt, J.F, Masui, R, Kuramitsu, S. | | Deposit date: | 2013-02-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins.
Febs Lett., 587, 2013
|
|
5GHK
 
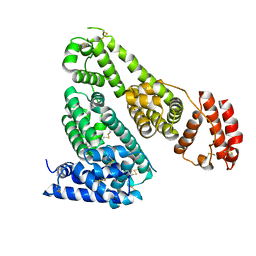 | | Crystal Structure Analysis of Canine serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yamada, K, Kureishi, M, Yokomaku, K, Shinohara, R, Akiyama, M, Komatsu, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Artificial Blood for Dogs
Sci Rep, 6, 2016
|
|
2ZDX
 
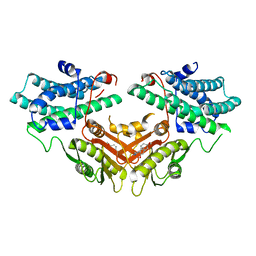 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3A8S
 
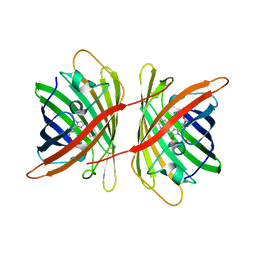 | |
3WU6
 
 | | Oxidized E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WT0
 
 | |
7WDO
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDR
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, Beta-glucosidase, SULFATE ION | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDS
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-xylopyranose | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7WDP
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
