3JCK
 
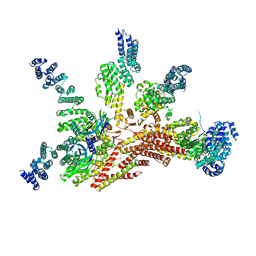 | | Structure of the yeast 26S proteasome lid sub-complex | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | Authors: | Herzik Jr, M.A, Dambacher, C.M, Worden, E.J, Martin, A, Lander, G.C. | | Deposit date: | 2015-12-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition.
Elife, 5, 2016
|
|
3HTE
 
 | | Crystal structure of nucleotide-free hexameric ClpX | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SULFATE ION | | Authors: | Glynn, S.E, Martin, A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.026 Å) | | Cite: | Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine.
Cell(Cambridge,Mass.), 139, 2009
|
|
7Q6L
 
 | |
7Q48
 
 | |
1AZ3
 
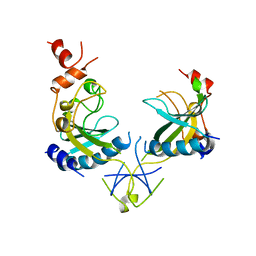 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1AZ4
 
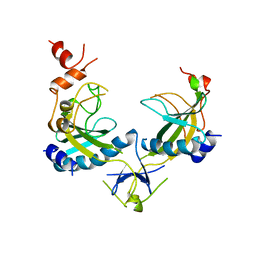 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B, T93A MUTANT | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
4O8Y
 
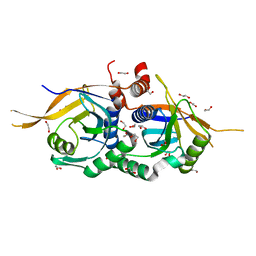 | | Zinc-free Rpn11 in complex with Rpn8 | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8 | | Authors: | Worden, E.J, Padovani, C, Martin, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2LZP
 
 | |
4O8X
 
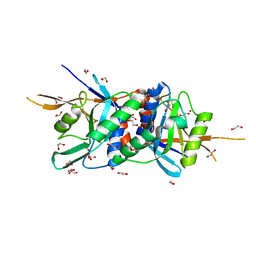 | | Zinc-bound Rpn11 in complex with Rpn8 | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8, ... | | Authors: | Worden, E.J, Padovani, C, Martin, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2MKB
 
 | |
2LZQ
 
 | |
5U4P
 
 | | Protein-protein complex between 26S proteasome regulatory subunit RPN8, RPN11, and Ubiquitin S31 | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8, Ubiquitin-40S ribosomal protein S31, ... | | Authors: | Worden, E.J, Dong, K.C, Martin, A. | | Deposit date: | 2016-12-05 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome.
Mol. Cell, 67, 2017
|
|
