5KCI
 
 | | Crystal Structure of HTC1 | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | Authors: | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | Deposit date: | 2016-06-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
7JLX
 
 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ (TIR domains) | | Descriptor: | Disease resistance protein Roq1 | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JLV
 
 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Disease resistance protein Roq1, MAGNESIUM ION | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7JLU
 
 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | CALCIUM ION, Disease resistance protein Roq1, XopQ | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
4IWR
 
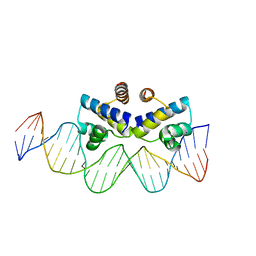 | | C.Esp1396I bound to a 25 base pair operator site | | Descriptor: | DNA (25-MER), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IVZ
 
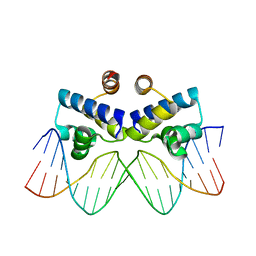 | |
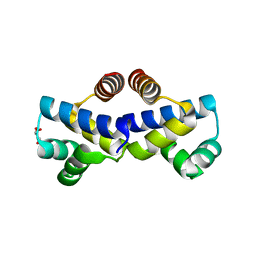
4FBI
 
 | |
4GLI
 
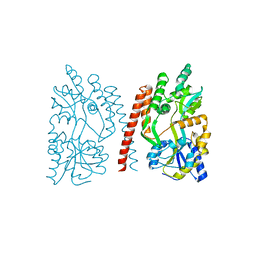 | | Crystal Structure of Human SMN YG-Dimer | | Descriptor: | Maltose-binding periplasmic protein, Survival motor neuron protein chimera | | Authors: | Martin, R.S, Perry, K, Van Duyne, G.D. | | Deposit date: | 2012-08-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The survival motor neuron protein forms soluble glycine zipper oligomers.
Structure, 20, 2012
|
|
4HAP
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I6R
 
 | |
4I6T
 
 | |
4I6U
 
 | | Crystal Structure of a Y37F mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-11-30 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Mutagenic Analysis of the RM Controller Protein C.Esp1396I.
Plos One, 9, 2014
|
|
4FN3
 
 | |
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8D
 
 | |
4I8T
 
 | | C.Esp1396I bound to a 19 base pair DNA duplex | | Descriptor: | DNA (5'-D(P*TP*GP*TP*GP*TP*GP*AP*TP*TP*AP*TP*AP*GP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*GP*AP*CP*TP*AP*TP*AP*AP*TP*CP*AP*CP*AP*CP*A)-3'), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-12-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IA8
 
 | |
5WO3
 
 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
447D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-18 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
4RG5
 
 | | Crystal Structure of S. Pombe SMN YG-Dimer | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein, Survival Motor Neuron protein chimera, ... | | Authors: | Gupta, K, Martin, R.S, Sarachan, K.L, Sharp, B, Van Duyne, G.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomeric Properties of Survival Motor NeuronGemin2 Complexes.
J.Biol.Chem., 290, 2015
|
|
966C
 
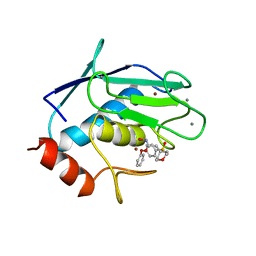 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
4YBA
 
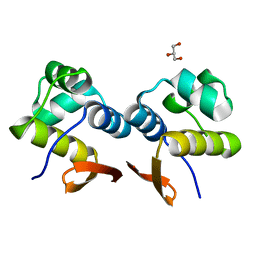 | | The structure of the C.Kpn2I controller protein | | Descriptor: | GLYCEROL, Regulatory protein C | | Authors: | Shevtsov, M.B, Martin, R.N, Swiderska, A, McGeehan, G.E, Kneale, G.G. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of the C.kpn2I controller protein
To Be Published
|
|
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | Descriptor: | Urotensin-2 receptor | | Authors: | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
