3GB8
 
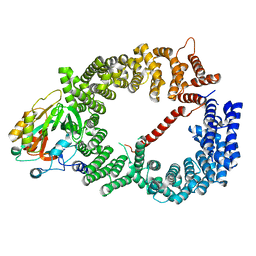 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
6HK4
 
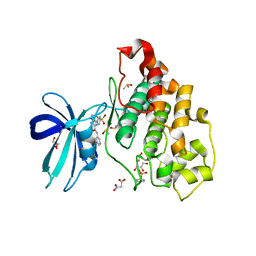 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | Descriptor: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK3
 
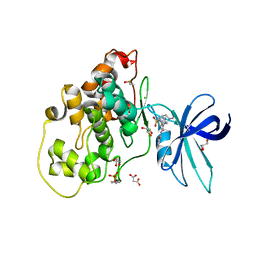 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6G5S
 
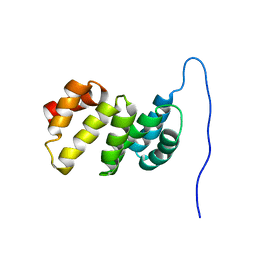 | | Solution structure of the TPR domain of the cell division coordinator, CpoB | | Descriptor: | Cell division coordinator CpoB | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Vollmer, W, Egan, A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6FZK
 
 | | NMR structure of UB2H, regulatory domain of PBP1b from E. coli | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Egan, A.J.F, Vollmer, W. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6G5R
 
 | |
2MMX
 
 | |
2MKW
 
 | |
2X4Z
 
 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
