1A7E
 
 | |
1A7D
 
 | |
4XNG
 
 | |
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
6QLY
 
 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | Descriptor: | E3 ubiquitin-protein ligase MYLIP | | Authors: | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
2MTG
 
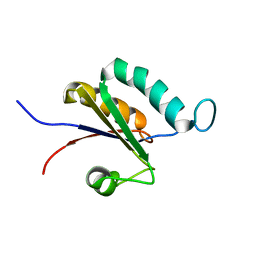 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2KFW
 
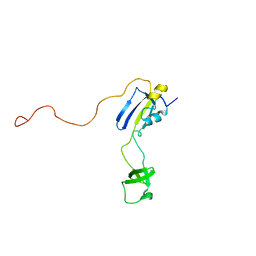 | | Solution structure of full-length SlyD from E.coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase slyD | | Authors: | Martino, L, He, Y, Hands-Taylor, K.L, Valentine, E.R, Kelly, G, Giancola, C, Conte, M.R. | | Deposit date: | 2009-02-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The interaction of the Escherichia coli protein SlyD with nickel ions illuminates the mechanism of regulation of its peptidyl-prolyl isomerase activity.
Febs J., 276, 2009
|
|
2JT7
 
 | |
1EEH
 
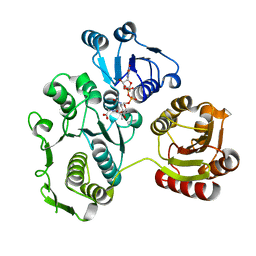 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
4UQP
 
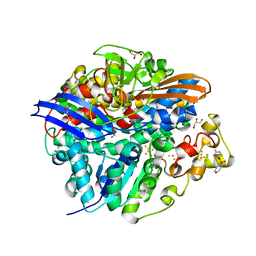 | | High-resolution structure of the D. fructosovorans NiFe-hydrogenase L122A mutant after exposure to air | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UQL
 
 | | High-resolution structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
3VDZ
 
 | | Tailoring Encodable Lanthanide-Binding Tags as MRI Contrast Agents: xq-dSE3-Ubiquitin at 2.4 Angstroms | | Descriptor: | GADOLINIUM ATOM, SULFATE ION, Ubiquitin-40S ribosomal protein S27a | | Authors: | Daughtry, K.D, Martin, L.J, Surraju, A, Imperiali, B, Allen, K.N. | | Deposit date: | 2012-01-06 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tailoring encodable lanthanide-binding tags as MRI contrast agents.
Chembiochem, 13, 2012
|
|
4UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
4UPV
 
 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UPE
 
 | | Structure of the unready Ni-A state of the S499C mutant of D. fructosovorans NiFe-hydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UD2
 
 | | Structure of anaerobically purified D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-05 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
4UD6
 
 | | Structure of methylviologen-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4UEQ
 
 | | Structure of the V74C large subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CALCIUM ION, CARBONATE ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4UE6
 
 | | Structure of methylene blue-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
4UEW
 
 | | Structure of H2-treated anaerobically purified D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
6Y1X
 
 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
3H3X
 
 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
6HTK
 
 | | X-ray structure of the tryptophan lyase NosL in complex with (R)-(+)-indoline-2-carboxylate | | Descriptor: | 1H-indole-2-carboxylic acid, 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Martin, L, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
