3ZF3
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84I mutant). | | Descriptor: | DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF0
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
4NDN
 
 | | Structural insights of MAT enzymes: MATa2b complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
3ZF5
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84F mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF4
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZEZ
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
2WE4
 
 | | Carbamate kinase from Enterococcus faecalis bound to a sulfate ion and two water molecules, which mimic the substrate carbamyl phosphate | | Descriptor: | CARBAMATE KINASE 1, SULFATE ION | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site-Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2WE5
 
 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | Authors: | Ramon-Maiques, S, Marina, A, Rubio, V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
3DGF
 
 | |
3DGE
 
 | |
7Q0N
 
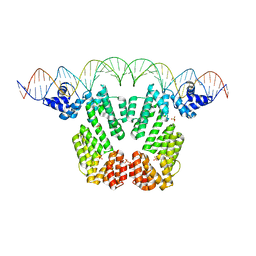 | | Arbitrium receptor from Katmira phage | | Descriptor: | Arbitrium receptor, DNA (45-MER), SULFATE ION | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of action of the arbitrium communication system in SPbeta phages.
Nat Commun, 13, 2022
|
|
7PWJ
 
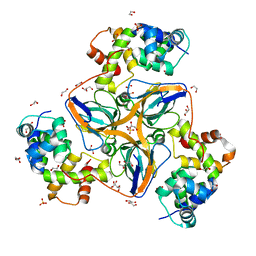 | | dUTPase from human in complex with Stl | | Descriptor: | 1,2-ETHANEDIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | Authors: | Sanz-Frasquet, C, Marina, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
5A19
 
 | | The structure of MAT2A in complex with PPNP. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5A1G
 
 | | The structure of Human MAT2A in complex with S-adenosylethionine and PPNP. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (DIPHOSPHONO)AMINOPHOSPHONIC ACID, IMIDAZOLE, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5A1I
 
 | | The structure of Human MAT2A in complex with SAM, Adenosine, Methionine and PPNP. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, ADENOSINE, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7ZUT
 
 | | Penicillium expansum chimera loop1 | | Descriptor: | Antifungal protein, CHLORIDE ION, SULFATE ION | | Authors: | Gallego, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZTJ
 
 | | Penicillium expansum antifungal protein chimera C-ter | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZVH
 
 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZW2
 
 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
7ZTF
 
 | | Penicillium expansum antifungal protein B | | Descriptor: | Antifungal protein, CHLORIDE ION, SODIUM ION | | Authors: | Gallego del Sol, F, Marina, A, Manzanares, P, Marcos, J.F, Giner Llorca, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rationally designed antifungal protein chimeras reveal new insights into structure-activity relationship.
Int.J.Biol.Macromol., 225, 2023
|
|
1GSJ
 
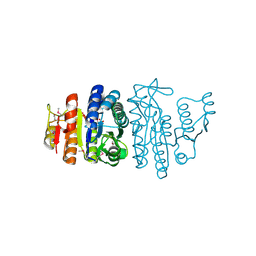 | | Selenomethionine substituted N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate n-acetyl-L-glutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
8ANT
 
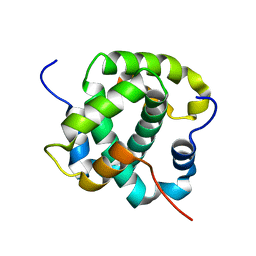 | |
8ANU
 
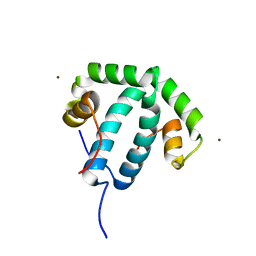 | | Crystal structure of protein phi3T-93 | | Descriptor: | NICKEL (II) ION, YopN. Phi3T_93 | | Authors: | Zamora-Caballero, S, Marina, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.310147 Å) | | Cite: | Antagonistic interactions between phage and host factors control arbitrium lysis-lysogeny decision.
Nat Microbiol, 9, 2024
|
|
8BJ6
 
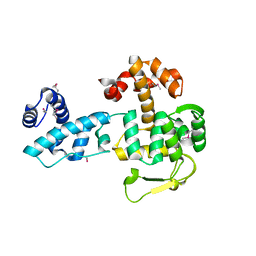 | | Crystal structure of YopR | | Descriptor: | SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a unique repression system present in arbitrium phages of the SPbeta family.
Cell Host Microbe, 31, 2023
|
|
8BJV
 
 | | Crystal structure of YopR | | Descriptor: | GLYCEROL, SPbeta prophage-derived uncharacterized protein YopR | | Authors: | Gallego del Sol, F, Marina, A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a unique repression system present in arbitrium phages of the SPbeta family.
Cell Host Microbe, 31, 2023
|
|
