1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
1DLJ
 
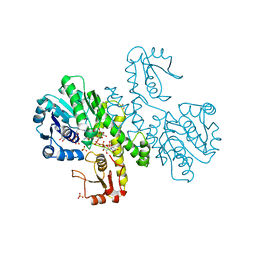 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1DLI
 
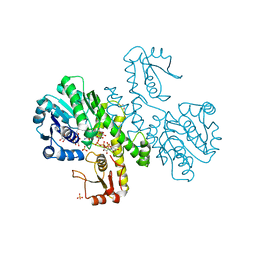 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1EH5
 
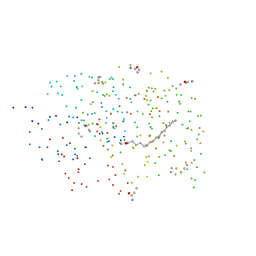 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 COMPLEXED WITH PALMITATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1RI3
 
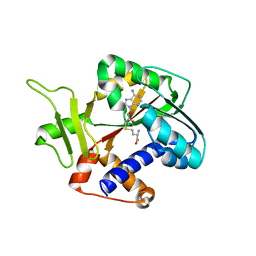 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
5NGV
 
 | |
1RI5
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
6X5J
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 2-(4-HYDROXY-5-PHENYL-1H-PYRAZOL-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.513 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X5L
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1RI1
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
6X5P
 
 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | Descriptor: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | Authors: | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1RI2
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
8OOR
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
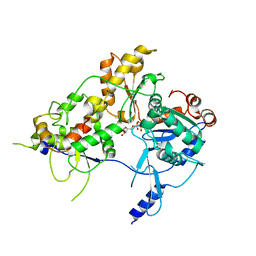 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOC
 
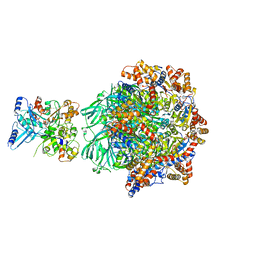 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
1EI9
 
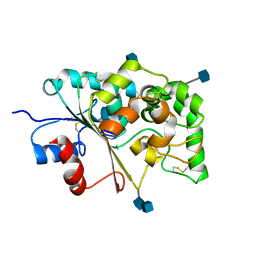 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOYL PROTEIN THIOESTERASE 1 | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1SNN
 
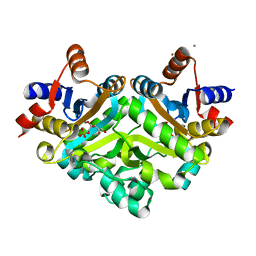 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
1TJO
 
 | | Iron-oxo clusters biomineralizing on protein surfaces. Structural analysis of H.salinarum DpsA in its low and high iron states | | Descriptor: | FE (III) ION, Iron-rich dpsA-homolog protein, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2004-06-07 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1K0W
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Strynadka, N.C.J. | | Deposit date: | 2001-09-21 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization
Biochemistry, 40, 2001
|
|
1H92
 
 | | SH3 domain of human Lck tyrosine kinase | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Schweimer, K, Hoffmann, S, Friedrich, U, Biesinger, B, Roesch, P, Sticht, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-10-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck
Biochemistry, 41, 2002
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
1FOF
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 | | Descriptor: | BETA LACTAMASE OXA-10, COBALT (II) ION, SULFATE ION | | Authors: | Paetzel, M, Danel, F, de Castro, L, Mosimann, S.C, Page, M.G.P, Strynadka, N.C.J. | | Deposit date: | 2000-08-28 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the class D beta-lactamase OXA-10.
Nat.Struct.Biol., 7, 2000
|
|
