2P6W
 
 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | Authors: | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
1USY
 
 | | ATP phosphoribosyl transferase (HisG:HisZ) complex from Thermotoga maritima | | Descriptor: | ATP PHOSPHORIBOSYLTRANSFERASE, ATP PHOSPHORIBOSYLTRANSFERASE REGULATORY SUBUNIT, HISTIDINE, ... | | Authors: | Vega, M.C, Fernandez, F.J, Murphy, G.E, Zou, P, Popov, A, Wilmanns, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-15 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Regulation of the Hetero-Octameric ATP Phosphoribosyl Transferase Complex from Thermotoga Maritima by a tRNA Synthetase-Like Subunit.
Mol.Microbiol., 55, 2005
|
|
3RBS
 
 | |
2FKK
 
 | | Crystal structure of the C-terminal domain of the bacteriophage T4 gene product 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2006-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Evolution of bacteriophage tails: structure of t4 gene product 10
J.Mol.Biol., 358, 2006
|
|
4GQ9
 
 | | Chikungunya virus neutralizing antibody 9.8B Fab fragment | | Descriptor: | Chikungunya virus neutralizing antibody 9.8B Fab fragment heavy chain, Chikungunya virus neutralizing antibody 9.8B Fab fragment light chain | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2012-08-22 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
1UU1
 
 | | Complex of Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER | | Authors: | Vega, M.C, Fernandez, F.J, Lehman, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
2FL9
 
 | |
1EL6
 
 | | STRUCTURE OF BACTERIOPHAGE T4 GENE PRODUCT 11, THE INTERFACE BETWEEN THE BASEPLATE AND SHORT TAIL FIBERS | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP11 | | Authors: | Leiman, P.G, Kostyuchenko, V.A, Schneider, M.M, Kurochkina, L.P, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2000-03-13 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of bacteriophage T4 gene product 11, the interface between the baseplate and short tail fibers.
J.Mol.Biol., 301, 2000
|
|
3H2T
 
 | | Crystal structure of gene product 6, baseplate protein of bacteriophage T4 | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-14 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of gene product 6 of bacteriophage t4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
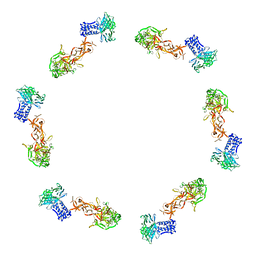
2FL8
 
 | |
3H3W
 
 | | Fitting of the gp6 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 dome-shaped baseplate | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
1M4J
 
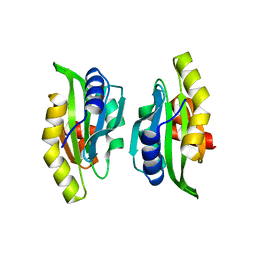 | | CRYSTAL STRUCTURE OF THE N-TERMINAL ADF-H DOMAIN OF MOUSE TWINFILIN ISOFORM-1 | | Descriptor: | A6 gene product | | Authors: | Paavilainen, V.O, Merckel, M.C, Falck, S, Ojala, P.J, Pohl, E, Wilmanns, M, Lappalainen, P. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation Between the Actin Monomer-binding Sites of Twinfilin and Actin-depolymerizing Factor (ADF)/Cofilin
J.Biol.Chem., 277, 2002
|
|
3H3Y
 
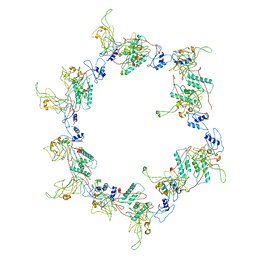 | | Fitting of the gp6 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 star-shaped baseplate | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
2L41
 
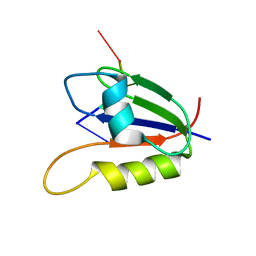 | | Nab3 RRM - UCUU complex | | Descriptor: | RNA (5'-R(P*UP*CP*UP*U)-3'), RRM domain from Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Stefl, R, Pergoli, R, Hobor, F, Kubicek, K, Zimmermann, M, Pasulka, J, Hofr, C. | | Deposit date: | 2010-09-28 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein
J.Biol.Chem., 286, 2011
|
|
1DGI
 
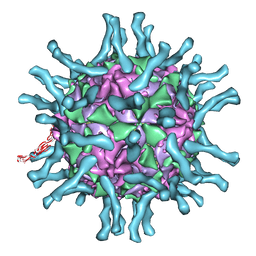 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3EPU
 
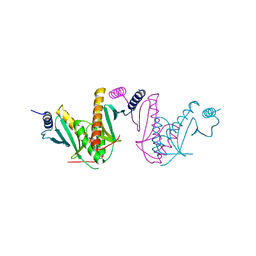 | | Crystal Structure of STM2138, a novel virulence chaperone in Salmonella | | Descriptor: | STM2138 Virulence Chaperone | | Authors: | Zhang, K, Andres, S.N, Hannemann, M, Coombes, B, Junop, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and quantitative proteomic
interactome of a novel virulence chaperone in Salmonella
To be Published
|
|
1EM9
 
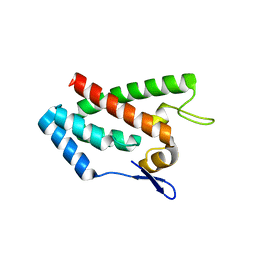 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: N-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27, MAGNESIUM ION | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1S0T
 
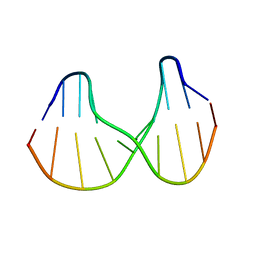 | | Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV | | Descriptor: | 5'-D(*Cp*Gp*Tp*Cp*Gp*Tp*Gp*Gp*Ap*C)-3', 5'-D(*Gp*Tp*Cp*Cp*(A3A)p*Cp*Gp*Ap*Cp*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-04 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
4HUD
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15. | | Descriptor: | Tail connector protein Gp15 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
8CQU
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1680) | | Descriptor: | CITRIC ACID, Putative NADH dehydrogenase/NAD(P)H nitroreductase, TERTIARY-BUTYL ALCOHOL | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQV
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQT
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1316) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Putative NADH dehydrogenase/NAD(P)H nitroreductase | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQS
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_0217) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like protein, PHOSPHATE ION | | Authors: | Blaha, J, Gratzl, S, Mortensen, S.A, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
5M7G
 
 | | Tubulin-MTD147 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(2,6-dimorpholin-4-ylpyridin-4-yl)-4-(trifluoromethyl)pyridin-2-amine, CALCIUM ION, ... | | Authors: | Bohnacker, T, Prota, A.E, Steinmetz, M.O, Wymann, M.P. | | Deposit date: | 2016-10-27 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
