2XIX
 
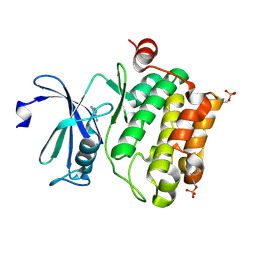 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
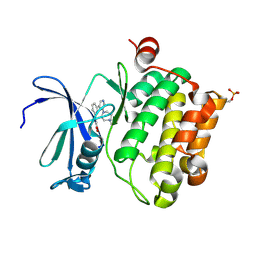 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIY
 
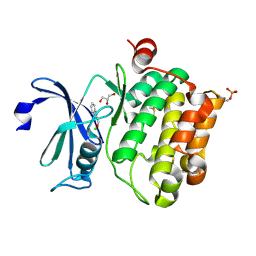 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5DPV
 
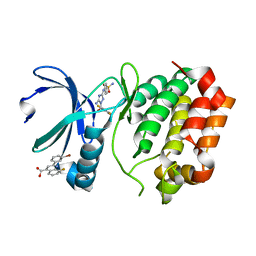 | | Aurora A Kinase in Complex with AA35 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DT0
 
 | | Aurora A Kinase in Complex with JNJ-7706621 in Space Group P6122 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DR9
 
 | | Aurora A Kinase in Complex with AA29 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-6-chloroquinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DN3
 
 | | Aurora A in complex with ATP and AA35. | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G, Huggins, D, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DR2
 
 | | Aurora A Kinase in Complex with AA30 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)quinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DT4
 
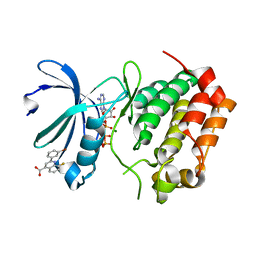 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
8G45
 
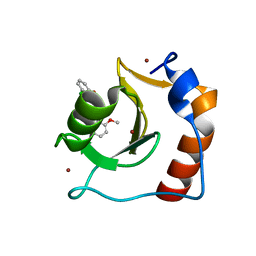 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
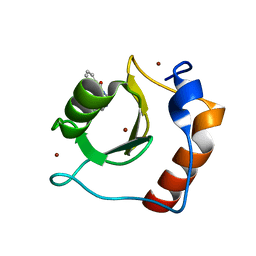 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
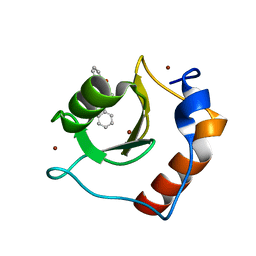 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
4UV0
 
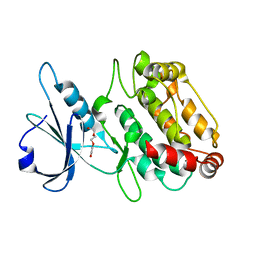 | | Structure of a semisynthetic phosphorylated DAPK | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, TRIETHYLENE GLYCOL | | Authors: | de Diego, I, Rios, P, Meyer, C, Koehn, M, Wilmanns, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular Mechanisms Behind Dapk Regulation: How the Phosphorylation Activity Switch Works
To be Published
|
|
2Y25
 
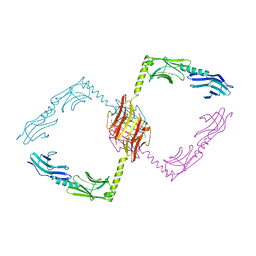 | |
2Y23
 
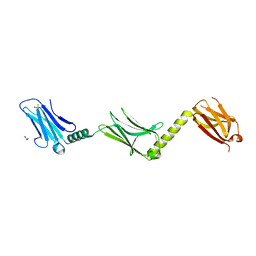 | |
2XQ1
 
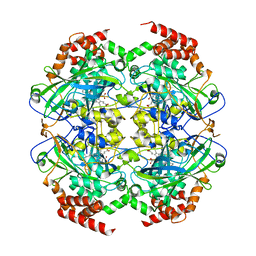 | | Crystal structure of peroxisomal catalase from the yeast Hansenula polymorpha | | Descriptor: | PEROXISOMAL CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Penya-Soler, E, Vega, M.C, Wilmanns, M, Williams, C.P. | | Deposit date: | 2010-08-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Features of Peroxisomal Catalase from the Yeast Hansenula Polymorpha
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5OCQ
 
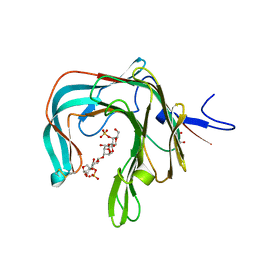 | | Crystal structure of the complex of the kappa-carrageenase from Pseudoalteromonas carrageenovora with an oligotetrasaccharide of kappa-carrageenan | | Descriptor: | 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, CITRIC ACID, ... | | Authors: | Czjzek, M, Leroux, C, Bernard, T, Matard-Mann, M, Jeudy, A, Michel, G. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
5OCR
 
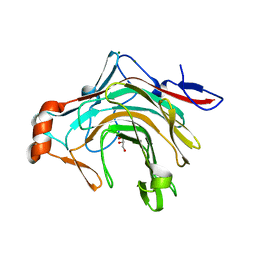 | | Crystal structure of the kappa-carrageenase zobellia_236 from Zobellia galactanivorans | | Descriptor: | GLYCEROL, Kappa-carrageenase, MAGNESIUM ION | | Authors: | Czjzek, M, Matard-Mann, M, Michel, G, Jeudy, A, Larocque, R. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
1VZW
 
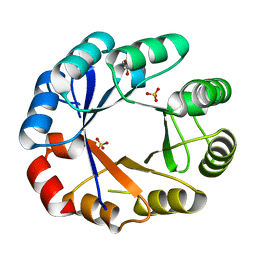 | | Crystal structure of the bifunctional protein Pria | | Descriptor: | GLYCEROL, PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Kuper, J, Wilmanns, M. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two-Fold Repeated (Beta-Alpha)(4) Half-Barrels May Provide a Molecular Tool for Dual Substrate Specificity
Embo Rep., 6, 2005
|
|
5K3V
 
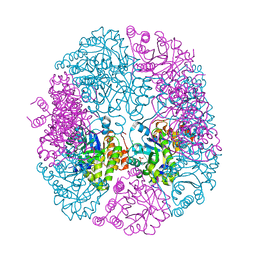 | | apo-PDX1.3 (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-20 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5W3M
 
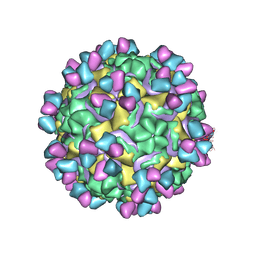 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:1, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KI7
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KI5
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIE
 
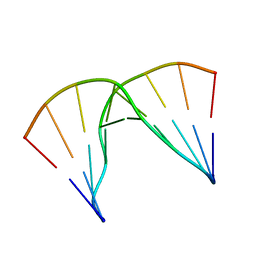 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*GP*GP*A)-R(P*G)-D(P*CP*TP*C)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
