1K9V
 
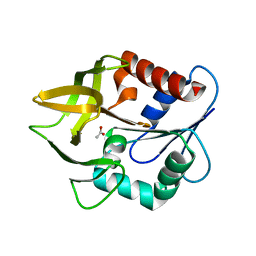 | | Structural evidence for ammonia tunelling across the (beta-alpha)8-barrel of the imidazole glycerol phosphate synthase bienzyme complex | | Descriptor: | ACETIC ACID, Amidotransferase hisH | | Authors: | Douangamath, A, Walker, M, Beismann-Driemeyer, S, Vega-Fernandez, M.C, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-10-31 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
Structure, 10, 2002
|
|
1NOH
 
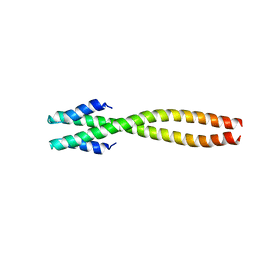 | | The structure of bacteriophage phi29 scaffolding protein gp7 after prohead assembly | | Descriptor: | HEAD MORPHOGENESIS PROTEIN | | Authors: | Morais, M.C, Kanamaru, S, Badasso, M.O, Koti, J.S, Owen, B.L, McMurray, C.T, L Anderson, D, Rossmann, M.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bacteriophage f29 scaffolding protein gp7 before and after prohead assembly
Nat.Struct.Biol., 10, 2003
|
|
1N5Z
 
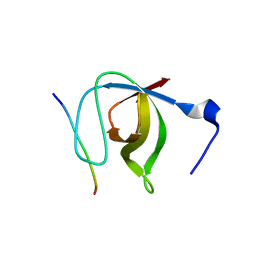 | | Complex structure of Pex13p SH3 domain with a peptide of Pex14p | | Descriptor: | 14-mer peptide from Peroxisomal membrane protein PEX14, Peroxisomal membrane protein PAS20 | | Authors: | Douangamath, A, Filipp, F.V, Klein, A.T.J, Barnett, P, Zou, P, Voorn-Brouwer, T, Vega, M.C, Mayans, O.M, Sattler, M, Distel, B, Wilmanns, M. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
MOL.CELL, 10, 2002
|
|
1N80
 
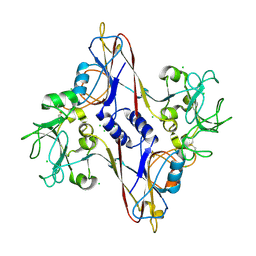 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1NA1
 
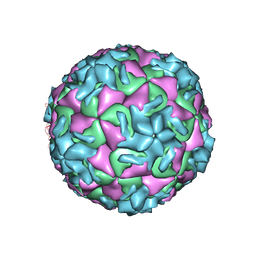 | | The structure of HRV14 when complexed with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Coat protein VP1, Coat protein VP2, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
4YWA
 
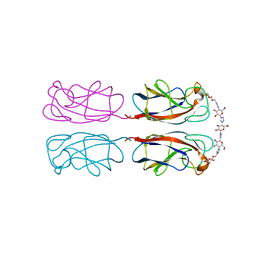 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | (2R,3R,4S,5R,6R,2'R,3'R,4'S,5'R,6'R)-2,2'-([(2R,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]bis{1H-1,2,3-triazole-1,4-diyl[(2S,3R,4S,5S,6S)-3,4-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2,5-diyl]-1H-1,2,3-triazole-1,4-diylmethanediyloxy})bis[6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol], CALCIUM ION, PA-I galactophilic lectin | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
1K28
 
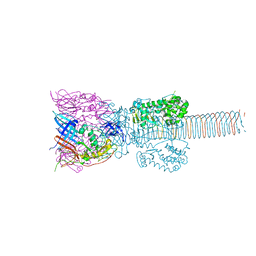 | | The Structure of the Bacteriophage T4 Cell-Puncturing Device | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP27, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kanamaru, S, Leiman, P.G, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Arisaka, F, Rossmann, M.G. | | Deposit date: | 2001-09-26 | | Release date: | 2002-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the cell-puncturing device of bacteriophage T4.
Nature, 415, 2002
|
|
2A08
 
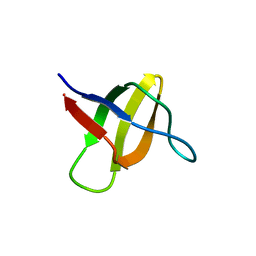 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
2R07
 
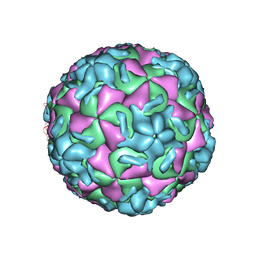 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(6-CHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R04
 
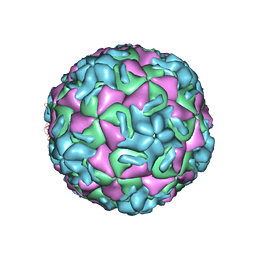 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R06
 
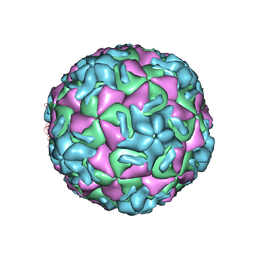 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2YKL
 
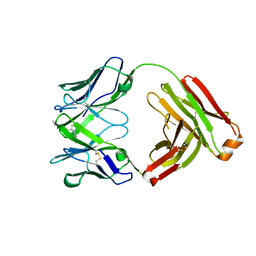 | | Structure of human anti-nicotine Fab fragment in complex with nicotine-11-yl-methyl-(4-ethylamino-4-oxo)-butanoate | | Descriptor: | FAB FRAGMENT, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Tars, K, Kotelovica, S, Lipowsky, G, Bauer, M, Beerli, R, Bachmann, M, Maurer, P. | | Deposit date: | 2011-05-27 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Binding Modes of Free and Carrier-Protein-Coupled Nicotine in a Human Monoclonal Antibody.
J.Mol.Biol., 415, 2012
|
|
6ZW0
 
 | |
6ZVZ
 
 | |
1OOT
 
 | |
1PDM
 
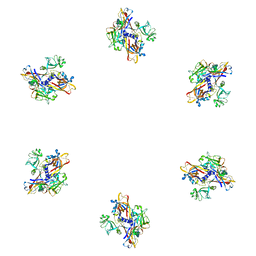 | | Fitting of gp8 structure into the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp8 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDP
 
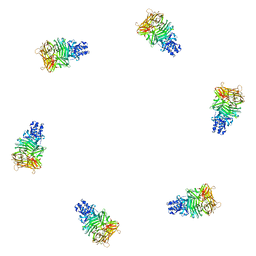 | | Fitting of gp9 structure into the bacteriophage T4 baseplate cryoEM reconstruction | | Descriptor: | Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
2VSX
 
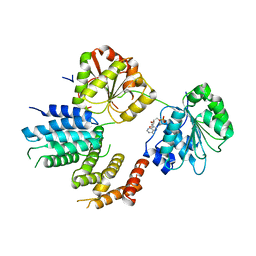 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-30 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VSO
 
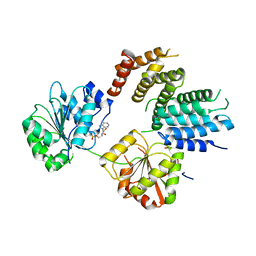 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3N7X
 
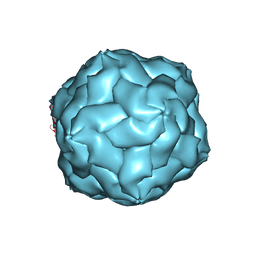 | |
3B3I
 
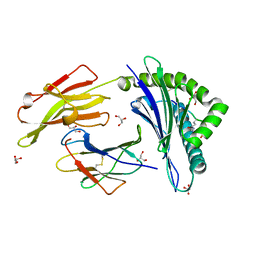 | | Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Paladini, F, Sorrentino, R, Saenger, W, Kumar, P, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes.
J.Biol.Chem., 283, 2008
|
|
1Z9Z
 
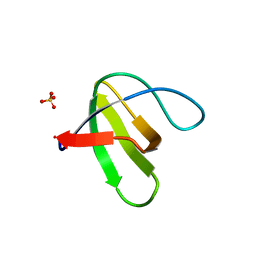 | | Crystal structure of yeast sla1 SH3 domain 3 | | Descriptor: | Cytoskeleton assembly control protein SLA1, SULFATE ION | | Authors: | Kursula, P, Kursula, I, Salmazo, A.P.T, Zou, P, Song, Y.H, Lehmann, F, Wilmanns, M. | | Deposit date: | 2005-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Yeast SH3 domain three-dimensional proteome
To be Published
|
|
7AYG
 
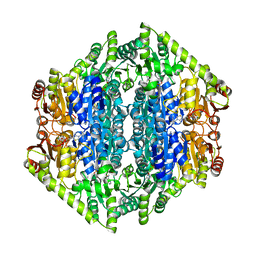 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
1ZUK
 
 | | Yeast BBC1 Sh3 domain complexed with a peptide from Las17 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Myosin tail region-interacting protein MTI1, ... | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-05-31 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
