5JXK
 
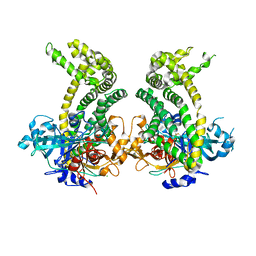 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JY0
 
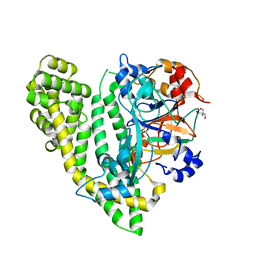 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JXP
 
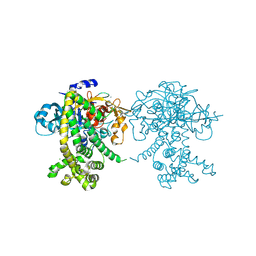 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5HID
 
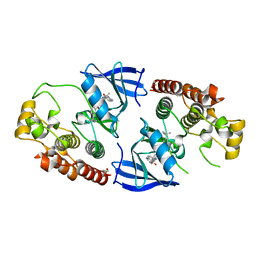 | | BRAF Kinase domain b3aC loop deletion mutant in complex with AZ628 | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HI2
 
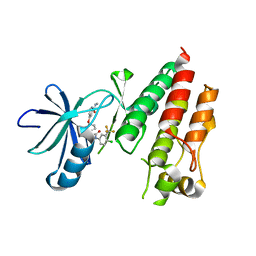 | | BRAF Kinase domain b3aC loop deletion mutant in complex with sorafenib | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HIE
 
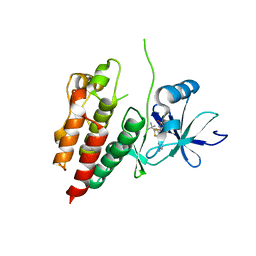 | | BRAF Kinase domain b3aC loop deletion mutant in complex with dabrafenib | | Descriptor: | Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
8POS
 
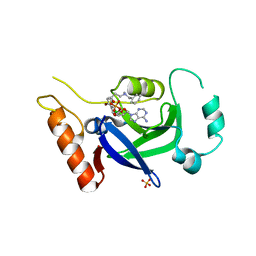 | | Crystal structure of wolbachia leucyl-tRNA synthetase editing domain bound to cmpd9-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
8POT
 
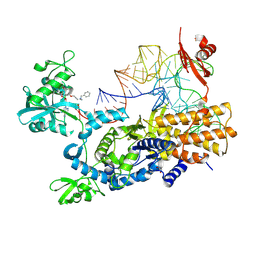 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole cmpd9 in the editing conformation | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate, ... | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
7NDY
 
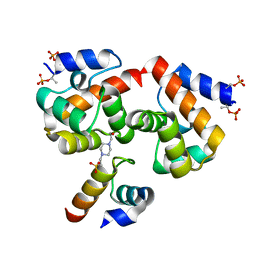 | | Di-phosphorylated Barrier-to-Autointegration Factor (BAF) in complex with LEM domain of Emerin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Barrier-to-autointegration factor, N-terminally processed, ... | | Authors: | Marcelot, A, Le Du, M.H, Hoffmann, G, Zinn-Justin, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
5M7N
 
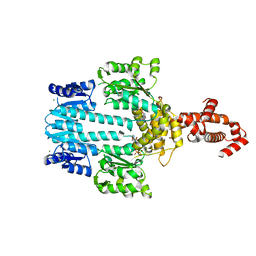 | | Crystal structure of NtrX from Brucella abortus in complex with ATP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5M7P
 
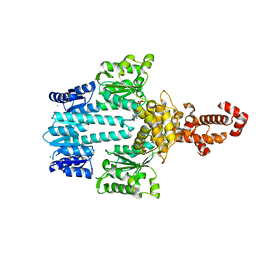 | | Crystal structure of NtrX from Brucella abortus in complex with ADP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5M7O
 
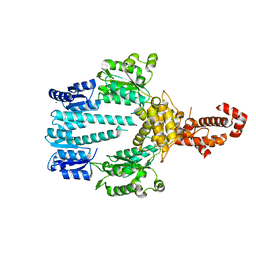 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
