2GQM
 
 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GQL
 
 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GQK
 
 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GVP
 
 | | Solution structure of Human apo Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Paulmaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT5
 
 | | Solution structure of apo Human Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT6
 
 | | Solution structure of Human Cu(I) Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1SP0
 
 | | Solution Structure of apoCox11 | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
6Z2Z
 
 | |
6Z2U
 
 | |
5EOE
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 (orthorhombic form) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
5EOO
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 (monoclinic form) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
5EUA
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 in complex with Moxalactam | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, Beta-lactamase, SODIUM ION | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
6TLG
 
 | | Ligand-free state of human 14-3-3 sigma isoform | | Descriptor: | 14-3-3 protein sigma, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
1Q3A
 
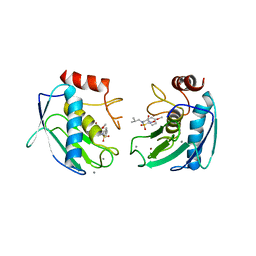 | | Crystal structure of the catalytic domain of human matrix metalloproteinase 10 | | Descriptor: | CALCIUM ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, Stromelysin-2, ... | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-07-29 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic domain of human matrix metalloproteinase 10.
J.Mol.Biol., 336, 2004
|
|
6ZRG
 
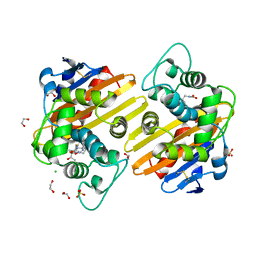 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed doripenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-({[amino(dihydroxy)methyl]amino}methyl)pyrrolidin-3-yl]sulfanyl}-5-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRJ
 
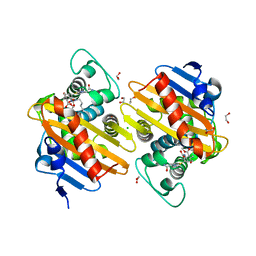 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRI
 
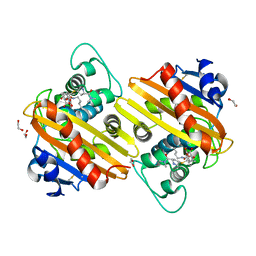 | | Crystal structure of OXA-10loop24 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRP
 
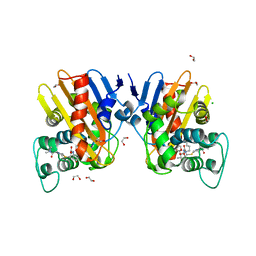 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, De Luca, F, Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S, Doquier, J.D. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRH
 
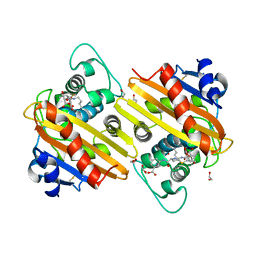 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZSX
 
 | |
6ZSW
 
 | |
6ZW2
 
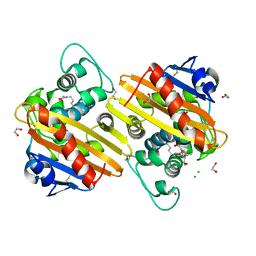 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6TBX
 
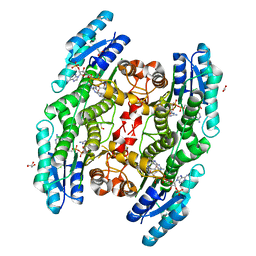 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with a tricyclic-based inhibitor | | Descriptor: | 2,4-bis(azanyl)-9~{H}-pyrimido[4,5-b]indol-6-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-11-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Trypanosoma brucei pteridine reductase 1 in complex with an innovative tricyclic-based inhibitor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TLF
 
 | | human 14-3-3 sigma isoform in complex with IMP | | Descriptor: | 14-3-3 protein sigma, INOSINIC ACID, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
