2V8O
 
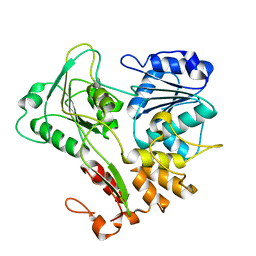 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
1W4B
 
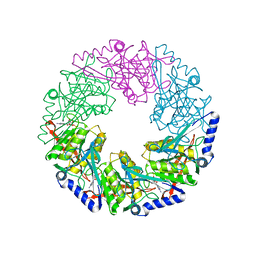 | | P4 protein from PHI12 in complex with product (AMPcPP Mg 22C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W4C
 
 | | P4 protein from Bacteriophage PHI12 apo state | | Descriptor: | NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W47
 
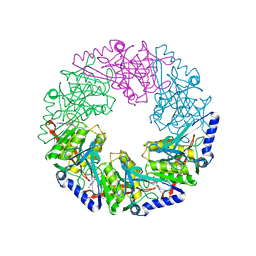 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W44
 
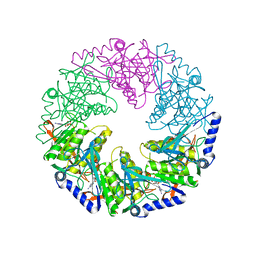 | | P4 protein from Bacteriophage PHI12 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W4A
 
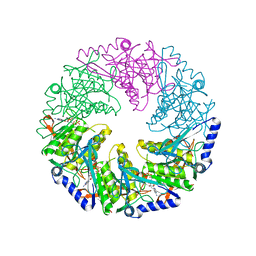 | | P4 protein from PHI12 in complex with AMPcPP and Mn | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W49
 
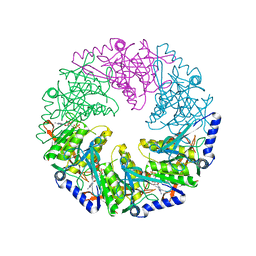 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W46
 
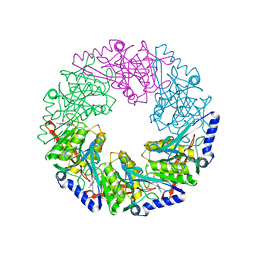 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W48
 
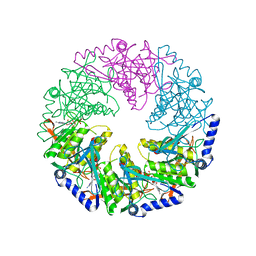 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1DYL
 
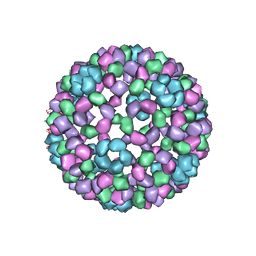 | | 9 ANGSTROM RESOLUTION CRYO-EM RECONSTRUCTION STRUCTURE OF SEMLIKI FOREST VIRUS (SFV) AND FITTING OF THE CAPSID PROTEIN STRUCTURE IN THE EM DENSITY | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Mancini, E.J, Clarke, M, Gowen, B.E, Rutten, T, Fuller, S.D. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Electron Microscopy Reveals the Functional Organization of an Enveloped Virus, Semliki Forest Virus.
Mol.Cell, 5, 2000
|
|
2JJX
 
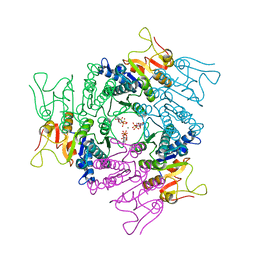 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
7OGS
 
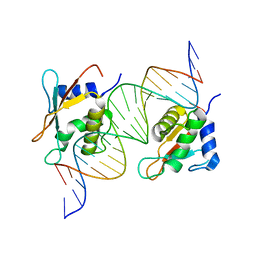 | |
7OOT
 
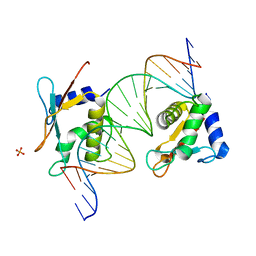 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element | | Descriptor: | DNA (5'-D(P*AP*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), DNA (5'-D(P*TP*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor 4, ... | | Authors: | Agnarelli, A, El Omari, K, Alt, A.O, Mancini, E.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to interferon-stimulated response element
To Be Published
|
|
2WV9
 
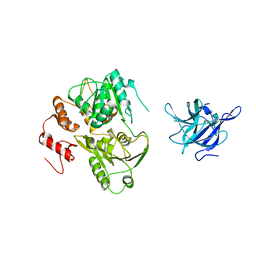 | | Crystal Structure of the NS3 protease-helicase from Murray Valley encephalitis virus | | Descriptor: | FLAVIVIRIN PROTEASE NS2B REGULATORY SUBUNIT, FLAVIVIRIN PROTEASE NS3 CATALYTIC SUBUNIT | | Authors: | Assenberg, R, Mastrangelo, E, Walter, T.S, Verma, A, Milani, M, Owens, R.J, Stuart, D.I, Grimes, J.M, Mancini, E.J. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Novel Conformational State of the Flavivirus Ns3 Protein: Implications for Polyprotein Processing and Viral Replication.
J.Virol., 83, 2009
|
|
6Q3M
 
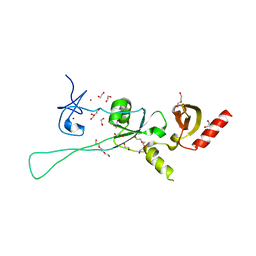 | | Structure of CHD4 PHD2 - tandem chromodomains | | Descriptor: | 1,2-ETHANEDIOL, Chromodomain-helicase-DNA-binding protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alt, A, Mancini, E.J. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of histone readers
To Be Published
|
|
5MOM
 
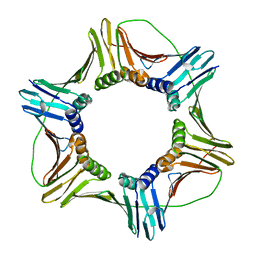 | |
6GUW
 
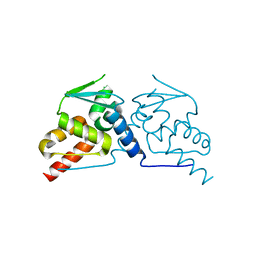 | |
6GUV
 
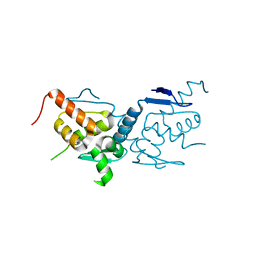 | | BTB domain of mouse PATZ1 | | Descriptor: | POZ (BTB) and AT hook-containing zinc finger 1 | | Authors: | Alt, A, Piepoli, S, Erman, B, Mancini, E.J. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis of the PATZ1 BTB domain homodimer
Acta Crystallogr.,Sect.D, 2020
|
|
6GUU
 
 | |
7O56
 
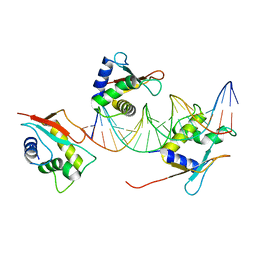 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element solved by Phosphorus and Sulphur SAD methods | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*AP*GP*AP*AP*AP*CP*CP*GP*AP*AP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*TP*TP*CP*GP*GP*TP*TP*TP*CP*TP*TP*TP*TP*AP*T)-3'), Interferon regulatory factor 4 | | Authors: | El Omari, K, Agnarelli, A, Duman, R, Wagner, A, Mancini, E.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-05-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
9F5B
 
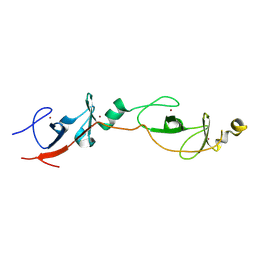 | | Identification of zinc ions in LMO4. | | Descriptor: | LIM domain transcription factor LMO4,LIM domain-binding protein 1, ZINC ION | | Authors: | El Omari, K, Forsyth, I, Mancini, E.J, Wagner, A. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Utilizing anomalous signals for element identification in macromolecular crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2VHC
 
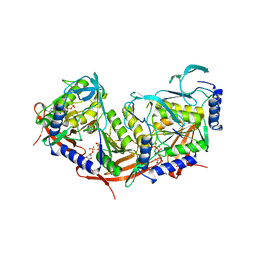 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHJ
 
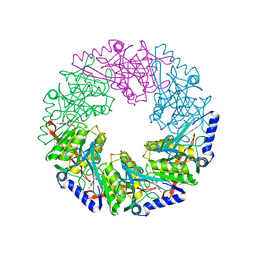 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHQ
 
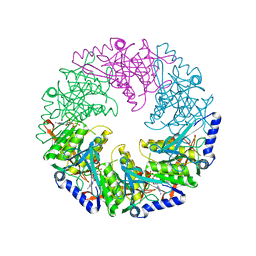 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ATP AND MG | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHT
 
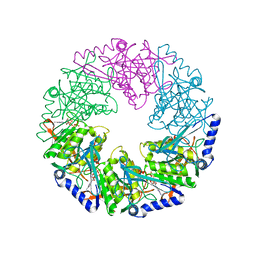 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 R279A mutant in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
