3KK8
 
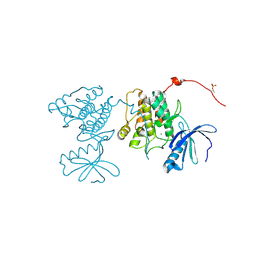 | | CaMKII Substrate Complex A | | Descriptor: | Calcium/calmodulin dependent protein kinase II, MAGNESIUM ION | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7MPS
 
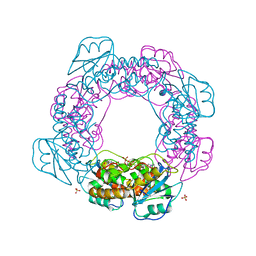 | | Brucella melitensis NrnC with engaged loop | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQD
 
 | | Bartonella henselae NrnC complexed with pAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPT
 
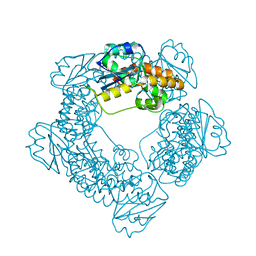 | | Brucella melitensis NrnC with bound Mg2+ | | Descriptor: | MAGNESIUM ION, NanoRNase C, PHOSPHATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQF
 
 | | Bartonella henselae NrnC complexed with pAAAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPU
 
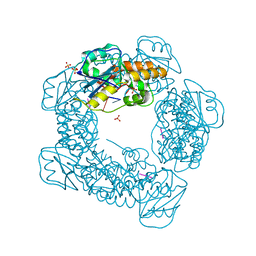 | | Brucella melitensis NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPP
 
 | |
7MQE
 
 | | Bartonella henselae NrnC complexed with pAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQB
 
 | | Bartonella henselae NrnC bound to pGG. D4 Symmetry | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQC
 
 | | Bartonella henselae NrnC bound to pGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPL
 
 | | Bartonella henselae NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQI
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQG
 
 | | Bartonella henselae NrnC complexed with pAAAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPM
 
 | | Bartonella henselae NrnC bound to pAA | | Descriptor: | 5'-phosphorylated AA, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPN
 
 | | Bartonella henselae NrnC bound to pGC | | Descriptor: | 5'-phosphorylated GC, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPQ
 
 | |
7MQH
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. D4 Symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
5C6L
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using intense Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
5C6J
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
7AQU
 
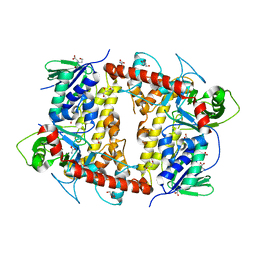 | |
7AQV
 
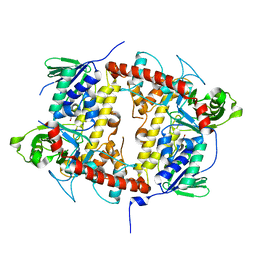 | |
2X8W
 
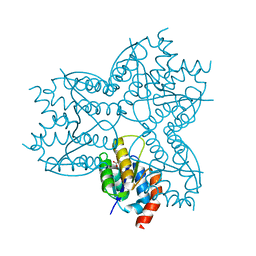 | | The Crystal Structure of Methylglyoxal Synthase from Thermus sp. GH5 Bound to Malonate. | | Descriptor: | MALONATE ION, METHYLGLYOXAL SYNTHASE | | Authors: | Shahsavar, A, Erfani Moghaddam, M, Antonyuk, S.V, Khajeh, K, Naderi-Manesh, H. | | Deposit date: | 2010-03-13 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Methylglyoxal Synthase from Thermus Sp.Gh5 in the Open and Closed Conformational States Provide Insight Into the Mechanism of Allosteric Regulation
To be Published
|
|
7PZD
 
 | | Cryo-EM structure of the NLRP3 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Hochheiser, I.V, Hagelueken, G, Behrmann, H, Behrmann, E, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Directionality of PYD filament growth determined by the transition of NLRP3 nucleation seeds to ASC elongation.
Sci Adv, 8, 2022
|
|
2CFD
 
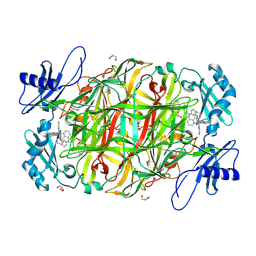 | | AGAO in complex with wc4l3 (Ru-wire inhibitor, 4-carbon linker, lambda enantiomer, data set 3) | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-20 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
7Z2K
 
 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
