7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD4
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase soaked with Cu+ ions | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD0
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Cd2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD1
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Hg2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD2
 
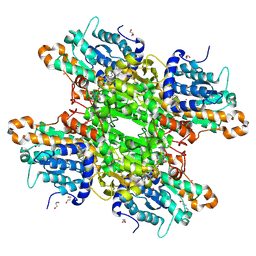 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Co2+ ions. | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
7ZD3
 
 | | Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Zn2+ ions | | Descriptor: | 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ADENOSINE, ... | | Authors: | Malecki, P.H, Gawel, M, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A closer look at molecular mechanisms underlying inhibition of S -adenosyl-L-homocysteine hydrolase by transition metal cations.
Chem.Commun.(Camb.), 60, 2024
|
|
6ETS
 
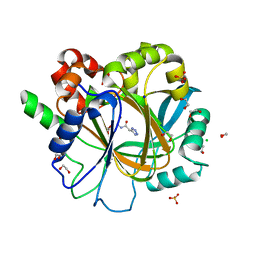 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F5R
 
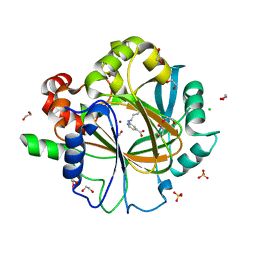 | | Crystal Structure of KDM4D with GF028 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-oxidanylpropylamino)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Crystal Structure of KDM4D with GF028 ligand
To be published
|
|
6ETT
 
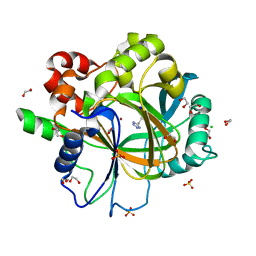 | | Crystal structure of KDM4D with tetrazole compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETV
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F5T
 
 | | Crystal Structure of KDM4D with tetrazole ligand GF057 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of KDM4D with tetrazole ligand
To be published
|
|
6H0Z
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067
To be published
|
|
6G9C
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis, TsES1 | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
6H11
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028
To be published
|
|
6H0X
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040
To be published
|
|
6F5Q
 
 | | Crystal Structure of KDM4D with GF026 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(methylsulfonylamino)ethylamino]pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of KDM4D with GF026 ligand
To be published
|
|
6ETG
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6G5W
 
 | | Crystal Structure of KDM4A with compound YP-03-038 | | Descriptor: | (4~{R})-5-methyl-4-phenyl-2-pyridin-2-yl-pyrazolidin-3-one, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
6G5X
 
 | | Crystal Structure of KDM4A with compound YP-02-145 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-methyl-5-oxidanylidene-4-phenyl-4~{H}-pyrazol-1-yl)-3~{H}-benzimidazole-5-carboxylic acid, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
6H0W
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035 | | Descriptor: | (2~{R})-3-phenyl-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035
To be published
|
|
6G9E
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis - TsES1 - 6 molecules in ASU | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
6H10
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073
To be published
|
|
6H0Y
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022 | | Descriptor: | (2~{R})-3-(4-methoxyphenyl)-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.212 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022
To be published
|
|
8CFC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment A01 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry A01
To be published
|
|
