1N4K
 
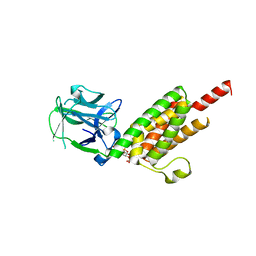 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2002-10-31 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
2N9C
 
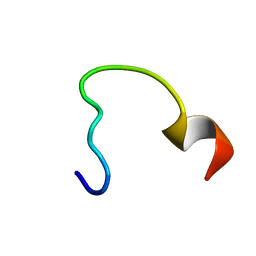 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
1NWD
 
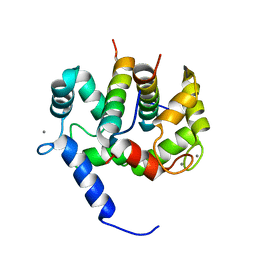 | | Solution Structure of Ca2+/Calmodulin bound to the C-terminal Domain of Petunia Glutamate Decarboxylase | | Descriptor: | CALCIUM ION, Calmodulin, Glutamate decarboxylase | | Authors: | Yap, K.L, Yuan, T, Mal, T.K, Vogel, H.J, Ikura, M. | | Deposit date: | 2003-02-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Simultaneous Binding of Two Carboxy-terminal Peptides
of Plant Glutamate Decarboxylase to Calmodulin
J.Mol.Biol., 328, 2003
|
|
