9F43
 
 | |
9F44
 
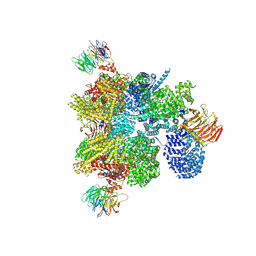 | |
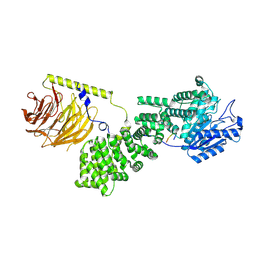
9F45
 
 | | cryo-EM structure of human LST2 bound to human mTOR complex 1 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Lateral signaling target protein 2 homolog, Regulatory-associated protein of mTOR, ... | | Authors: | Craigie, L.M, Maier, T. | | Deposit date: | 2024-04-26 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
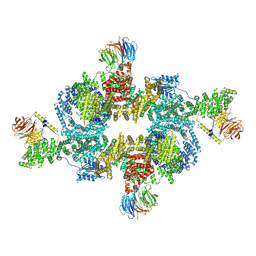
1W2B
 
 | | Trigger Factor ribosome binding domain in complex with 50S | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L13P, ... | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1W26
 
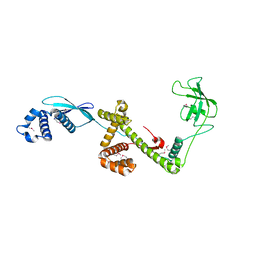 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | Descriptor: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | Authors: | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
3MF2
 
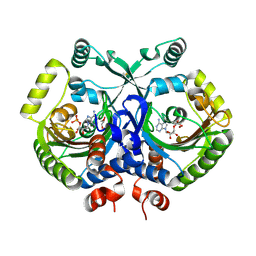 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bll0957 protein, GLYCEROL, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MF1
 
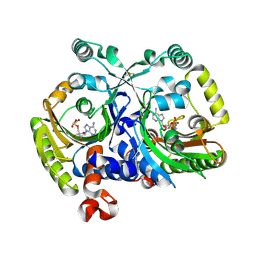 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with an analogue of glycyl adenylate | | Descriptor: | 5'-O-(glycylsulfamoyl)adenosine, Bll0957 protein, ZINC ION | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MEY
 
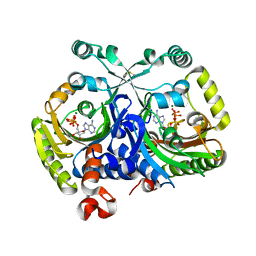 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bll0957 protein, MAGNESIUM ION, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2Z9A
 
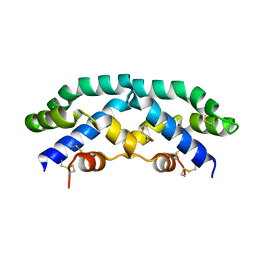 | |
6QGX
 
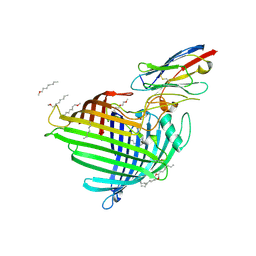 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6HD8
 
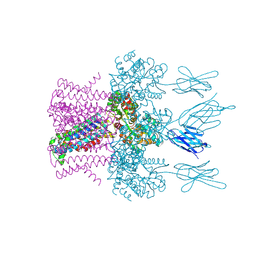 | | Crystal structure of the potassium channel MtTMEM175 in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDB
 
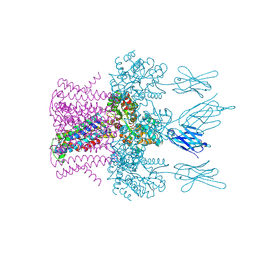 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HD9
 
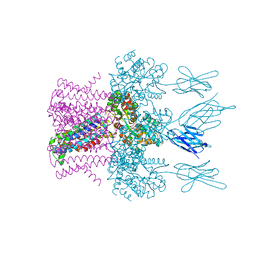 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDC
 
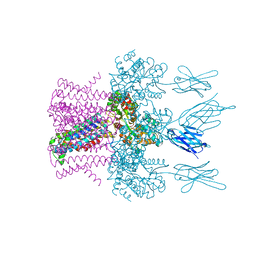 | | Crystal structure of the potassium channel MtTMEM175 T38A variant in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDA
 
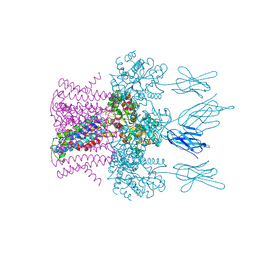 | | Crystal structure of the potassium channel MtTMEM175 with cesium | | Descriptor: | CESIUM ION, DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
5MUC
 
 | | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, Protein FimH | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Chemistry, 24, 2018
|
|
4TM8
 
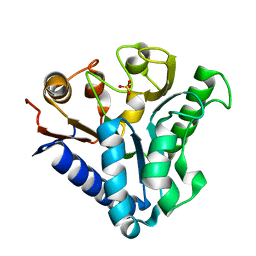 | |
4TM7
 
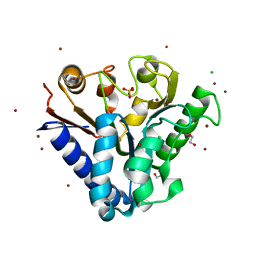 | | Crystal structure of 6-phosphogluconolactonase from Mycobacterium smegmatis N131D mutant soaked with CuSO4 | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphogluconolactonase, CHLORIDE ION, ... | | Authors: | Fujieda, N, Stuttfeld, E, Maier, T. | | Deposit date: | 2014-05-31 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Enzyme repurposing of a hydrolase as an emergent peroxidase upon metal binding.
Chem Sci, 6, 2015
|
|
7ZM9
 
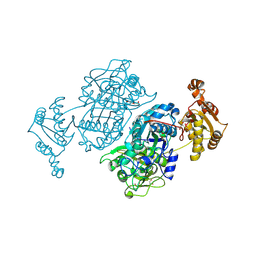 | | Ketosynthase domain 3 of Brevibacillus Brevis orphan BGC11 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMF
 
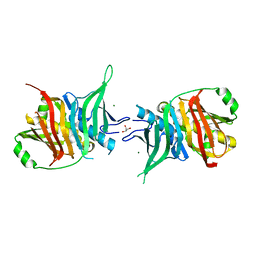 | | Dehydratase domain of module 3 from Brevibacillus Brevis orphan BGC11 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
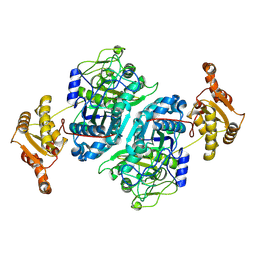
7ZMD
 
 | |
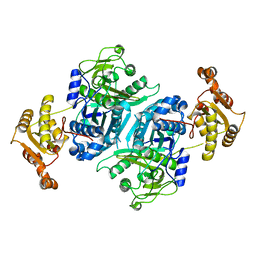
7ZMC
 
 | |
