6XAB
 
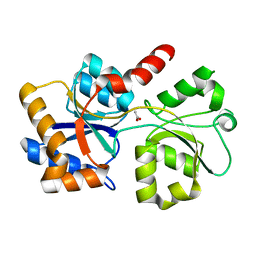 | |
6XAD
 
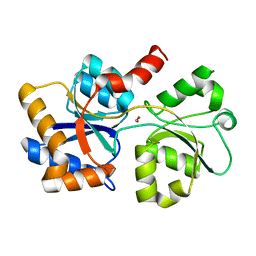 | |
6VDA
 
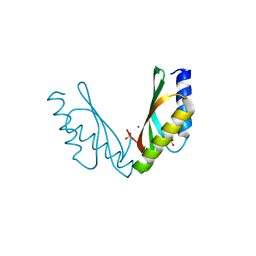 | |
6VD9
 
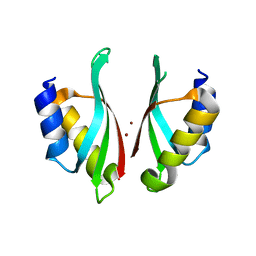 | |
6VD8
 
 | |
7L22
 
 | |
7KYP
 
 | |
7KYO
 
 | |
7MQZ
 
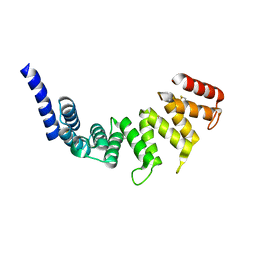 | | Cytochrome c oxidase assembly factor 7 | | Descriptor: | Cytochrome c oxidase assembly factor 7 | | Authors: | Maghool, S, Maher, M.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mitochondrial COA7 is a heme-binding protein with disulfide reductase activity, which acts in the early stages of complex IV assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3LX5
 
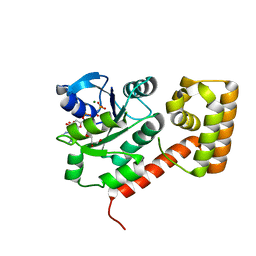 | | Crystal structure of mGMPPNP-bound NFeoB from S. thermophilus | | Descriptor: | 2-amino-9-(5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-3-O-{[2-(methylamino)phenyl]carbonyl}-beta-D-erythro-pentofuranosyl-2-ulose)-1,9-dihydro-6H-purin-6-one, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
5VDE
 
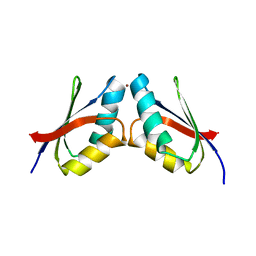 | |
1XGE
 
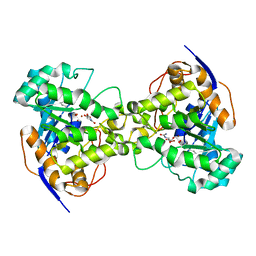 | | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between subunits | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Chan, C.W, Guss, J.M, Christopherson, R.I, Maher, M.J. | | Deposit date: | 2004-09-17 | | Release date: | 2005-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between Subunits
J.Mol.Biol., 348, 2005
|
|
7URH
 
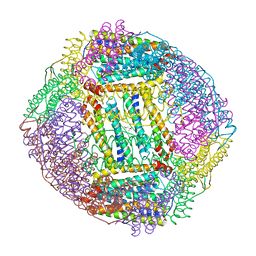 | | Crystal structure of Ferritin 2 from Caenorhabditis elegans, FTN-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, Ferritin | | Authors: | Malcolm, T.R, Maher, M.J, Mubarak, S.S.M. | | Deposit date: | 2022-04-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Biochemical Characterization of Caenorhabditis elegans Ferritins.
Biochemistry, 62, 2023
|
|
7USN
 
 | | Crystal structure of ferritin 1 from Caenorhabditis elegans, FTN-1 | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL, ... | | Authors: | Malcolm, T.R, Maher, M.J, Mubarak, S.S.M. | | Deposit date: | 2022-04-25 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Biochemical Characterization of Caenorhabditis elegans Ferritins.
Biochemistry, 62, 2023
|
|
3DSP
 
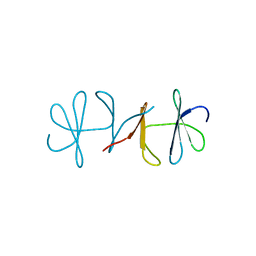 | | Crystal structure of apo copper resistance protein CopK | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Ash, M.-R, Maher, M.J. | | Deposit date: | 2008-07-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unprecedented binding cooperativity between Cu(I) and Cu(II) in the copper resistance protein CopK from Cupriavidus metallidurans CH34: implications from structural studies by NMR spectroscopy and X-ray crystallography
J.Am.Chem.Soc., 131, 2009
|
|
3DSO
 
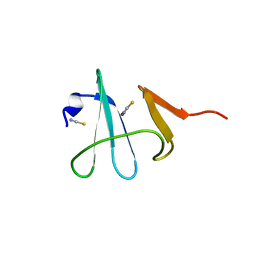 | | Crystal structure of Cu(I) bound copper resistance protein CopK | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein copK, THIOCYANATE ION | | Authors: | Ash, M.-R, Maher, M.J. | | Deposit date: | 2008-07-13 | | Release date: | 2009-03-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unprecedented binding cooperativity between Cu(I) and Cu(II) in the copper resistance protein CopK from Cupriavidus metallidurans CH34: implications from structural studies by NMR spectroscopy and X-ray crystallography
J.Am.Chem.Soc., 131, 2009
|
|
5VDF
 
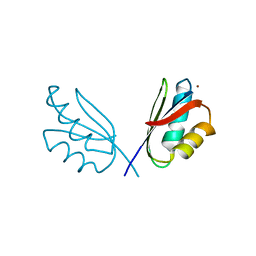 | |
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
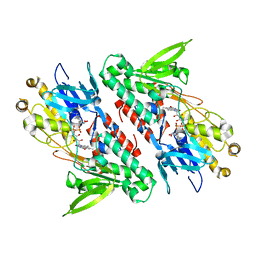 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
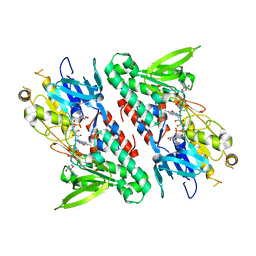 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3SS8
 
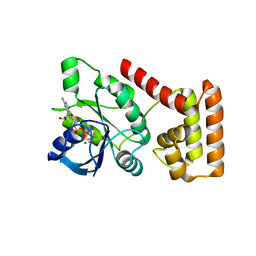 | | Crystal structure of NFeoB from S. thermophilus bound to GDP.AlF4- and K+ | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure
Plos One, 6, 2011
|
|
1RUT
 
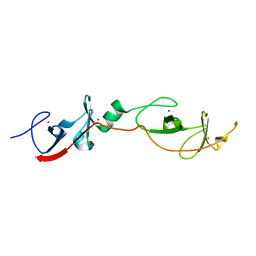 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
3TAH
 
 | | Crystal structure of an S. thermophilus NFeoB N11A mutant bound to mGDP | | Descriptor: | 3'-O-(N-methylanthraniloyl)guanosine-5'-diphosphate, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-08-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an N11A mutant of the G-protein domain of FeoB
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3TU6
 
 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
4PW9
 
 | |
